Potent SARS-CoV-2 neutralizing antibodies with therapeutic effects in two animal models
- PMID: 36406861
- PMCID: PMC9664764
- DOI: 10.1016/j.isci.2022.105596
Potent SARS-CoV-2 neutralizing antibodies with therapeutic effects in two animal models
Abstract
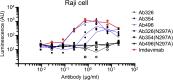
The use of therapeutic neutralizing antibodies against SARS-CoV-2 infection has been highly effective. However, there remain few practical antibodies against viruses that are acquiring mutations. In this study, we created 494 monoclonal antibodies from patients with COVID-19-convalescent, and identified antibodies that exhibited the comparable neutralizing ability to clinically used antibodies in the neutralization assay using pseudovirus and authentic virus including variants of concerns. These antibodies have different profiles against various mutations, which were confirmed by cell-based assay and cryo-electron microscopy. To prevent antibody-dependent enhancement, N297A modification was introduced. Our antibodies showed a reduction of lung viral RNAs by therapeutic administration in a hamster model. In addition, an antibody cocktail consisting of three antibodies was also administered therapeutically to a macaque model, which resulted in reduced viral titers of swabs and lungs and reduced lung tissue damage scores. These results showed that our antibodies have sufficient antiviral activity as therapeutic candidates.
Keywords: Unology; immune response; virology.
© 2022 The Author(s).
Conflict of interest statement
M.T., K.S., H.S., T.T., Y.T., S.M., H.F., M.S., T.M., K.K., Y.I., H.I., M.N., Y.Kitagawa, and Y.Kawaoka declared that they are co-inventors on a patent application on neutralizing antibodies described in this article (PCT/JP2021/35159). The remaining authors have no declarations of interest.
Figures






References
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases Infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Earnest R., Uddin R., Matluk N., Renzette N., Siddle K.J., Loreth C., Adams G., Tomkins-Tinch C.H., Petrone M.E., Rothman J.E., et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. medRxiv. 2021 doi: 10.1101/2021.10.06.21264641. Preprint at. - DOI - PMC - PubMed
-
- Thomas S.J., Moreira E.D., Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMoa2110345. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

