A sample-to-answer COVID-19 diagnostic device based on immiscible filtration and CRISPR-Cas12a-assisted detection
- PMID: 36406953
- PMCID: PMC9640297
- DOI: 10.1016/j.talo.2022.100166
A sample-to-answer COVID-19 diagnostic device based on immiscible filtration and CRISPR-Cas12a-assisted detection
Abstract
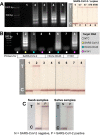
In response to the ongoing coronavirus disease 2019 (COVID-19) pandemic and disparities of vaccination coverage in low-and middle-income countries, it is vital to adopt a widespread testing and screening programme, combined with contact tracing, to monitor and effectively control the infection dispersion in areas where medical resources are limited. This work presents a lab-on-a-chip device, namely 'IFAST-LAMP-CRISPR', as an affordable, rapid and high-precision molecular diagnostic means for detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The herein proposed 'sample-to-answer' platform integrates RNA extraction, amplification and molecular detection with lateral flow readout in one device. The microscale dimensions of the device containing immiscible liquids, coupled with the use of silica paramagnetic beads and guanidine hydrochloride, streamline sample preparation (including RNA extraction, concentration and purification) in 15 min with minimal hands-on steps. The pre-amplification in combination with CRISPR-Cas12a detection assays targeting the nucleoprotein (N) gene achieved visual identification of ≥ 470 copies mL-1 genomic SARS-CoV-2 samples in 45 min. On-chip assays showed the ability to isolate and detect SARS-CoV-2 RNA from 100 genome copies mL-1 of replication-deficient viral particles in 1 h. This simple, affordable and integrated platform demonstrated a visual, faster, and yet specificity- and sensitivity-comparable alternative to the costly gold-standard reverse transcription-polymerase chain reaction (RT-PCR) assay, requiring only a simple heating source. Initial testing illustrates the platform viability both on nasopharyngeal swab and saliva samples collected using the easily accessible Swan-brand cigarette filter, providing a complete workflow for COVID-19 diagnostics in low-resource settings.
Keywords: COVID-19; CRISPR-Cas; Diagnostics; Immiscible filtration; Point-of-care; SARS-CoV-2.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article
Figures





References
-
- Coronavirus Resource Centre, Coronavirus Resource Center. John Hopkins University., https://coronavirus.jhu.edu/map.html.
-
- O.W.I. data, Coronavirus (COVID-19) vaccinations, 2021.
-
- W.H. Organization, 2022.
LinkOut - more resources
Full Text Sources
Miscellaneous
