WINNER: A network biology tool for biomolecular characterization and prioritization
- PMID: 36407327
- PMCID: PMC9672476
- DOI: 10.3389/fdata.2022.1016606
WINNER: A network biology tool for biomolecular characterization and prioritization
Abstract
Background and contribution: In network biology, molecular functions can be characterized by network-based inference, or "guilt-by-associations." PageRank-like tools have been applied in the study of biomolecular interaction networks to obtain further the relative significance of all molecules in the network. However, there is a great deal of inherent noise in widely accessible data sets for gene-to-gene associations or protein-protein interactions. How to develop robust tests to expand, filter, and rank molecular entities in disease-specific networks remains an ad hoc data analysis process.
Results: We describe a new biomolecular characterization and prioritization tool called Weighted In-Network Node Expansion and Ranking (WINNER). It takes the input of any molecular interaction network data and generates an optionally expanded network with all the nodes ranked according to their relevance to one another in the network. To help users assess the robustness of results, WINNER provides two different types of statistics. The first type is a node-expansion p-value, which helps evaluate the statistical significance of adding "non-seed" molecules to the original biomolecular interaction network consisting of "seed" molecules and molecular interactions. The second type is a node-ranking p-value, which helps evaluate the relative statistical significance of the contribution of each node to the overall network architecture. We validated the robustness of WINNER in ranking top molecules by spiking noises in several network permutation experiments. We have found that node degree-preservation randomization of the gene network produced normally distributed ranking scores, which outperform those made with other gene network randomization techniques. Furthermore, we validated that a more significant proportion of the WINNER-ranked genes was associated with disease biology than existing methods such as PageRank. We demonstrated the performance of WINNER with a few case studies, including Alzheimer's disease, breast cancer, myocardial infarctions, and Triple negative breast cancer (TNBC). In all these case studies, the expanded and top-ranked genes identified by WINNER reveal disease biology more significantly than those identified by other gene prioritizing software tools, including Ingenuity Pathway Analysis (IPA) and DiAMOND.
Conclusion: WINNER ranking strongly correlates to other ranking methods when the network covers sufficient node and edge information, indicating a high network quality. WINNER users can use this new tool to robustly evaluate a list of candidate genes, proteins, or metabolites produced from high-throughput biology experiments, as long as there is available gene/protein/metabolic network information.
Keywords: gene prioritization; network biology; network expansion; network statistical analysis; pathway analysis.
Copyright © 2022 Nguyen, Yue, Slominski, Welner, Zhang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


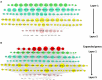

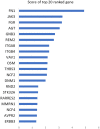


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

