Expression of alternative transcription factor 4 mRNAs and protein isoforms in the developing and adult rodent and human tissues
- PMID: 36407762
- PMCID: PMC9666405
- DOI: 10.3389/fnmol.2022.1033224
Expression of alternative transcription factor 4 mRNAs and protein isoforms in the developing and adult rodent and human tissues
Abstract
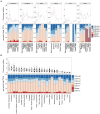
Transcription factor 4 (TCF4) belongs to the class I basic helix-loop-helix family of transcription factors (also known as E-proteins) and is vital for the development of the nervous system. Aberrations in the TCF4 gene are associated with several neurocognitive disorders such as schizophrenia, intellectual disability, post-traumatic stress disorder, depression, and Pitt-Hopkins Syndrome, a rare but severe autism spectrum disorder. Expression of the human TCF4 gene can produce at least 18 N-terminally distinct protein isoforms, which activate transcription with different activities and thus may vary in their function during development. We used long-read RNA-sequencing and western blot analysis combined with the analysis of publicly available short-read RNA-sequencing data to describe both the mRNA and protein expression of the many distinct TCF4 isoforms in rodent and human neural and nonneural tissues. We show that TCF4 mRNA and protein expression is much higher in the rodent brain compared to nonneural tissues. TCF4 protein expression is highest in the rodent cerebral cortex and hippocampus, where expression peaks around birth, and in the rodent cerebellum, where expression peaks about a week after birth. In human, highest TCF4 expression levels were seen in the developing brain, although some nonneural tissues displayed comparable expression levels to adult brain. In addition, we show for the first time that out of the many possible TCF4 isoforms, the main TCF4 isoforms expressed in the rodent and human brain and other tissues are TCF4-B, -C, -D, -A, and-I. Taken together, our isoform specific analysis of TCF4 expression in different tissues could be used for the generation of gene therapy applications for patients with TCF4-associated diseases.
Keywords: basic helix–loop–helix transcription factor; brain tissue; long-read RNA sequencing; neurodevelopment; peripheral tissue; transcription factor TCF4; western blot analysis.
Copyright © 2022 Sirp, Shubina, Tuvikene, Tamberg, Kiir, Kranich and Timmusk.
Conflict of interest statement
JT and TT were employed by Protobios LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Amiel J., Rio M., de Pontual L., Redon R., Malan V., Boddaert N., et al. . (2007). Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 80, 988–993. doi: 10.1086/515582, PMID: - DOI - PMC - PubMed
-
- Badowska D. M., Brzózka M. M., Kannaiyan N., Thomas C., Dibaj P., Chowdhury A., et al. . (2020). Modulation of cognition and neuronal plasticity in gain-and loss-of-function mouse models of the schizophrenia risk gene Tcf4. Transl. Psychiatry 10:343. doi: 10.1038/s41398-020-01026-7, PMID: - DOI - PMC - PubMed
-
- Brockschmidt A., Todt U., Ryu S., Hoischen A., Landwehr C., Birnbaum S., et al. . (2007). Severe mental retardation with breathing abnormalities (Pitt–Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum. Mol. Genet. 16, 1488–1494. doi: 10.1093/hmg/ddm099, PMID: - DOI - PubMed
-
- Brzózka M. M., Radyushkin K., Wichert S. P., Ehrenreich H., Rossner M. J. (2010). Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol. Psychiatry 68, 33–40. doi: 10.1016/j.biopsych.2010.03.015, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

