RNA biomarkers for alcohol use disorder
- PMID: 36407766
- PMCID: PMC9673015
- DOI: 10.3389/fnmol.2022.1032362
RNA biomarkers for alcohol use disorder
Abstract
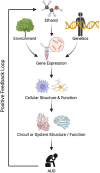
Alcohol use disorder (AUD) is highly prevalent and one of the leading causes of disability in the US and around the world. There are some molecular biomarkers of heavy alcohol use and liver damage which can suggest AUD, but these are lacking in sensitivity and specificity. AUD treatment involves psychosocial interventions and medications for managing alcohol withdrawal, assisting in abstinence and reduced drinking (naltrexone, acamprosate, disulfiram, and some off-label medications), and treating comorbid psychiatric conditions (e.g., depression and anxiety). It has been suggested that various patient groups within the heterogeneous AUD population would respond more favorably to specific treatment approaches. For example, there is some evidence that so-called reward-drinkers respond better to naltrexone than acamprosate. However, there are currently no objective molecular markers to separate patients into optimal treatment groups or any markers of treatment response. Objective molecular biomarkers could aid in AUD diagnosis and patient stratification, which could personalize treatment and improve outcomes through more targeted interventions. Biomarkers of treatment response could also improve AUD management and treatment development. Systems biology considers complex diseases and emergent behaviors as the outcome of interactions and crosstalk between biomolecular networks. A systems approach that uses transcriptomic (or other -omic data, e.g., methylome, proteome, metabolome) can capture genetic and environmental factors associated with AUD and potentially provide sensitive, specific, and objective biomarkers to guide patient stratification, prognosis of treatment response or relapse, and predict optimal treatments. This Review describes and highlights state-of-the-art research on employing transcriptomic data and artificial intelligence (AI) methods to serve as molecular biomarkers with the goal of improving the clinical management of AUD. Considerations about future directions are also discussed.
Keywords: RNA seq; alcohol dependence; alcoholism; biomarker; microarray; transcriptome.
Copyright © 2022 Ferguson, Mayfield and Messing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
New Australian guidelines for the treatment of alcohol problems: an overview of recommendations.Med J Aust. 2021 Oct 4;215 Suppl 7:S3-S32. doi: 10.5694/mja2.51254. Med J Aust. 2021. PMID: 34601742
-
Real-world effectiveness of pharmacological treatments of alcohol use disorders in a Swedish nation-wide cohort of 125 556 patients.Addiction. 2021 Aug;116(8):1990-1998. doi: 10.1111/add.15384. Epub 2021 Jan 14. Addiction. 2021. PMID: 33394527 Free PMC article.
-
Alcohol Use Disorder: The Role of Medication in Recovery.Alcohol Res. 2021 Jun 3;41(1):07. doi: 10.35946/arcr.v41.1.07. eCollection 2021. Alcohol Res. 2021. PMID: 34113531 Free PMC article. Review.
-
Perspectives on the pharmacological management of alcohol use disorder: Are the approved medications effective?Eur J Intern Med. 2022 Sep;103:13-22. doi: 10.1016/j.ejim.2022.05.016. Epub 2022 May 18. Eur J Intern Med. 2022. PMID: 35597734 Review.
-
Medications for treating alcohol use disorder: A narrative review.Alcohol Clin Exp Res (Hoboken). 2023 Jul;47(7):1224-1237. doi: 10.1111/acer.15118. Epub 2023 Jun 8. Alcohol Clin Exp Res (Hoboken). 2023. PMID: 37526592 Review.
Cited by
-
Next-generation biomarkers for alcohol consumption and alcohol use disorder diagnosis, prognosis, and treatment: A critical review.Alcohol Clin Exp Res (Hoboken). 2025 Jan;49(1):5-24. doi: 10.1111/acer.15476. Epub 2024 Nov 12. Alcohol Clin Exp Res (Hoboken). 2025. PMID: 39532676 Review.
References
-
- American Psychiatric Association [APA] (2013). Dsm-5 task force. Diagnostic and statistical manual of mental disorders: DSM-5. Arlington, VA: American Psychiatric Association. 10.1176/appi.books.9780890425596 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

