Autophagic lysosome reformation in health and disease
- PMID: 36409033
- PMCID: PMC10240999
- DOI: 10.1080/15548627.2022.2128019
Autophagic lysosome reformation in health and disease
Abstract
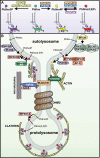
Lysosomes are the primary degradative compartment within cells and there have been significant advances over the past decade toward understanding how lysosome homeostasis is maintained. Lysosome repopulation ensures sustained autophagy function, a fundamental process that protects against disease. During macroautophagy/autophagy, cellular debris is sequestered into phagophores that mature into autophagosomes, which then fuse with lysosomes to generate autolysosomes in which contents are degraded. Autophagy cannot proceed without the sufficient generation of lysosomes, and this can be achieved via their de novo biogenesis. Alternatively, during autophagic lysosome reformation (ALR), lysosomes are generated via the recycling of autolysosome membranes. During this process, autolysosomes undergo significant membrane remodeling and scission to generate membrane fragments, that mature into functional lysosomes. By utilizing membranes already formed during autophagy, this facilitates an efficient pathway for re-deriving lysosomes, particularly under conditions of prolonged autophagic flux. ALR dysfunction is emerging as an important disease mechanism including for neurodegenerative disorders such as hereditary spastic paraplegia and Parkinson disease, neuropathies including Charcot-Marie-Tooth disease, lysosome storage disorders, muscular dystrophy, metabolic syndrome, and inflammatory and liver disorders. Here, we provide a comprehensive review of ALR, including an overview of its dynamic spatiotemporal regulation by MTOR and phosphoinositides, and the role ALR dysfunction plays in many diseases.
Keywords: Autophagic lysosome reformation; MTOR; PtdIns(4,5)P2; PtdIns4P; lysosome; phosphoinositide.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. 2020. Oct 15; 383(16):1564–1576. PubMed PMID: 33053285. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
