Global Phylogeny and F Virulence Plasmid Carriage in Pandemic Escherichia coli ST1193
- PMID: 36409140
- PMCID: PMC9769970
- DOI: 10.1128/spectrum.02554-22
Global Phylogeny and F Virulence Plasmid Carriage in Pandemic Escherichia coli ST1193
Abstract
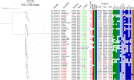
Lower urinary tract, renal, and bloodstream infections caused by phylogroup B2 extraintestinal pathogenic Escherichia coli (ExPEC) are a leading cause of morbidity and mortality. ST1193 is a phylogroup B2, multidrug-resistant sequence type that has risen to prominence globally, but a comprehensive analysis of the F virulence plasmids it carries is lacking. We performed a phylogenomic analysis of ST1193 (n = 707) whole-genome sequences from EnteroBase using entries with comprehensive isolation metadata. The data set comprised isolates from humans (n = 634 [90%]), including 339 (48%) from extraintestinal infection sites, and isolates from companion animals, wastewater, and wildlife. Phylogenetic analyses combined with gene detection and genotyping resolved an ST1193 clade structure segregated by serotype and F plasmid carriage. Most F plasmids fell into one of three related plasmid subtypes: F-:A1:B10 (n = 444 [65.97%]), F-:A1:B1 (n = 84 [12.48%]), and F-:A1:B20 (n = 80 [11.89%]), all of which carry the virulence genes cjrABC colocalized with senB (cjrABC-senB), a trademark signature of F29:A-:B10 subtype plasmids (pUTI89). To examine the phylogenetic relationship of these plasmids with pUTI89, complete sequences of F-:A1:B1 and F-:1:B20 plasmids were resolved. Unlike pUTI89, the most dominant and widely disseminated F plasmid that carries cjrABC-senB, F plasmids in ST1193 often carry a complex resistance region with an integron truncation (intI1Δ745) signature embedded within a structure assembled by IS26. Plasmid analysis shows that ST1193 has F plasmids that carry cjrABC-senB and ARG-encoding genes but lack tra regions and are likely derivatives of pUTI89. Further epidemiological investigation of ST1193 should seek to confirm its presence in human-associated environments and identify any potential agricultural links, which are currently lacking. IMPORTANCE We have generated an updated ST1193 phylogeny using publicly available sequences, reinforcing previous assertions that Escherichia coli ST1193 is a human-associated lineage, with many examples sourced from human extraintestinal infections. ST1193 from urban-adapted birds, wastewater, and companion animals are frequent, but isolates from animal agriculture are notably absent. Phylogenomic analysis identified several clades segregated by serogroup, all noted to carry highly similar F plasmids and antimicrobial resistance (AMR) signatures. Investigation of these plasmids revealed virulence regions with similarity to pUTI89, a key F virulence plasmid among dominant pandemic extraintestinal pathogenic E. coli lineages, and encoding a complex antibiotic resistance structure mobilized by IS26. This work has uncovered a series of F virulence plasmids in ST1193 and shows that the lineage mimics the host range and virulence attributes of other E. coli strains that carry pUTI89. These observations have significant ramifications for epidemiological source tracking of emerging and established pandemic ExPEC lineages.
Keywords: Escherichia coli; antimicrobial resistance; integron; pandemic; plasmids; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Johnson JR, Johnston BD, Porter SB, Clabots C, Bender TL, Thuras P, Trott DJ, Cobbold R, Mollinger J, Ferrieri P, Drawz S, Banerjee R. 2019. Rapid emergence, subsidence, and molecular eetection of Escherichia coli sequence type 1193-fimH64, a new disseminated multidrug-resistant commensal and extraintestinal pathogen. J Clin Microbiol 57:e01664-18. doi: 10.1128/JCM.01664-18. - DOI - PMC - PubMed
-
- Johnson TJ, Elnekave E, Miller EA, Munoz-Aguayo J, Flores Figueroa C, Johnston B, Nielson DW, Logue CM, Johnson JR. 2019. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother 63:e01913-18. doi: 10.1128/AAC.01913-18. - DOI - PMC - PubMed
-
- Tchesnokova V, Radey M, Chattopadhyay S, Larson L, Weaver JL, Kisiela D, Sokurenko EV. 2019. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc Natl Acad Sci USA 116:14740–14748. doi: 10.1073/pnas.1903002116. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

