Metagenomic Next-Generation Sequencing for the Diagnosis of Neonatal Infectious Diseases
- PMID: 36409152
- PMCID: PMC9769891
- DOI: 10.1128/spectrum.01195-22
Metagenomic Next-Generation Sequencing for the Diagnosis of Neonatal Infectious Diseases
Abstract
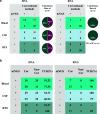
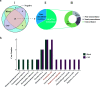
Infectious diseases pose a fatal risk to neonates. Timely and accurate pathogen detection is crucial for proper clinical diagnosis and therapeutic strategies. Limited sample volumes from neonatal patients seriously hindered the accurate detection of pathogens. Here, we unravel that metagenomic next-generation sequencing (mNGS) of cell-free DNA (cfDNA) and RNA can achieve unbiased detection of trace pathogens from different kinds of body fluid samples and blood samples. We enrolled 168 neonatal patients with suspected infections from whom blood samples (n = 153), cerebrospinal fluid samples (n = 127), and respiratory tract samples (RTSs) (including bronchoalveolar lavage fluids, sputa, and respiratory secretions) (n = 51) were collected and analyzed using mNGS. High rates of positivity (70.2%; 118/168) of mNGS were observed, and the coincidence rate against the final clinical diagnosis in positive mNGS cases reached 68.6% (81/118). The most common causative pathogens were Klebsiella pneumoniae (n = 12), Escherichia coli (n = 12), and Streptococcus pneumoniae (n = 8). mNGS using cfDNA and RNA can identify microbes that cannot be detected by conventional methods in different body fluid and blood samples, and more than 50% of these microbes were identified as causative pathogens. Further local polynomial regression fitting analysis revealed that the best timing for mNGS detection ranged from 1 to 3 days after the start of continuous antimicrobial therapy. Diagnosed and guided by mNGS results, the therapeutic regimens for 86 out of 117 neonatal patients were changed, most of whom (80/86) completely recovered and were discharged, while 44 out of 86 patients completely or partially stopped unnecessary medication. Our findings highlight the importance of mNGS in detecting causative DNA and RNA pathogens in infected neonatal patients. IMPORTANCE To the best of our knowledge, this is the first report on evaluating the performance of mNGS using cfDNA and RNA from body fluid and blood samples for diagnosing neonatal infections. mNGS of RNA and cfDNA can achieve the unbiased detection and identification of trace pathogens from different kinds of neonatal body fluid and blood samples with a high total coincidence rate (226/331; 68.3%) against final clinical diagnoses by sample. The best timing for mNGS detection in neonatal infections ranged from 1 to 3 days, rather than 0 days, after the start of continuous antimicrobial therapy. Our findings highlight the importance of mNGS in detecting causative DNA and RNA pathogens, and the extensive application of mNGS for the diagnosis of neonatal infections can be expected.
Keywords: antimicrobial therapy; infection; mNGS; neonate; the best timing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Seale AC, Blencowe H, Manu AA, Nair H, Bahl R, Qazi SA, Zaidi AK, Berkley JA, Cousens SN, Lawn JE, pSBI Investigator Group . 2014. Estimates of possible severe bacterial infection in neonates in sub-Saharan Africa, South Asia, and Latin America for 2012: a systematic review and meta-analysis. Lancet Infect Dis 14:731–741. doi: 10.1016/S1473-3099(14)70804-7. - DOI - PMC - PubMed
-
- Berhane M, Gidi NW, Eshetu B, Gashaw M, Tesfaw G, Wieser A, Bårnes GK, Froeschl G, Ali S, Gudina EK. 2021. Clinical profile of neonates admitted with sepsis to neonatal intensive care unit of Jimma Medical Center, a tertiary hospital in Ethiopia. Ethiop J Health Sci 31:485–494. doi: 10.4314/ejhs.v31i3.5. - DOI - PMC - PubMed
-
- Liu L, Oza S, Hogan D, Chu Y, Perin J, Zhu J, Lawn JE, Cousens S, Mathers C, Black RE. 2016. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the sustainable development goals. Lancet 388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. - DOI - PMC - PubMed
-
- Yadav M, Kumar M, Tripathi S, Singh SN. 2019. Tracheal aspirate in the diagnosis of neonatal pneumonia soon after birth: a prospective observational study. East Afr Sch J Med Sci 2:296–301.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

