Genetic characterization of a novel pheasant-origin orthoreovirus using Next-Generation Sequencing
- PMID: 36409667
- PMCID: PMC9678273
- DOI: 10.1371/journal.pone.0277411
Genetic characterization of a novel pheasant-origin orthoreovirus using Next-Generation Sequencing
Abstract
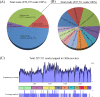
A field isolate (Reo/SDWF /Pheasant/17608/20) of avian orthoreovirus (ARV), isolated from a flock of game-pheasants in Weifang, Shandong Province, was genetically characterized being a field variant or novel strain in our recent research studies in conducting whole genome sequencing by using Next-Generation Sequencing (NGS) technique on Illumina MiSeq platform. Among a total of 870,197 35-151-mer sequencing reads, 297,711 reads (34.21%) were identified as ARV sequences. The de novo assembly of the ARV reads resulted in generation of 10 ARV-related contigs with the average sequencing coverage from 1390× to 1977× according to 10 ARV genome segments. The complete genomes of this pheasant-origin ARV (Reo/SDWF /Pheasant/17608/20) were 23,495 bp in length and consist of 10 dsRNA segments ranged from 1192 bp (S4) to 3958 bp (L1) encoding 12 viral proteins. Sequence comparison between the SDWF17608 and classic ARV reference strains revealed that 58.1-100% nucleotide (nt) identities and 51.4-100% amino acid (aa) identities were in genome segment coding genes. The 10 RNA segments had conversed termini at 5' (5'-GCUUUU) and 3' (UCAUC-3') side, which were identical to the most published ARV strains. Phylogenetic analysis revealed that this pheasant ARV field variant was closely related with chicken ARV strains in 7 genome segment genes, but it possessed significant sequence divergence in M1, M3 and S2 segments. These findings suggested that this pheasant-origin field variant was a divergent ARV strain and was likely originated from reassortments between different chicken ARV strains.
Copyright: © 2022 Tang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Detection and characterization of two co-infection variant strains of avian orthoreovirus (ARV) in young layer chickens using next-generation sequencing (NGS).Sci Rep. 2016 Apr 19;6:24519. doi: 10.1038/srep24519. Sci Rep. 2016. PMID: 27089943 Free PMC article.
-
Isolation and genomic characterization of a novel chicken-orign orthoreovirus causing goslings hepatitis.Vet Microbiol. 2018 Dec;227:69-77. doi: 10.1016/j.vetmic.2018.10.017. Epub 2018 Oct 25. Vet Microbiol. 2018. PMID: 30473354
-
Genomic characterization of a novel avian arthritis orthoreovirus variant by next-generation sequencing.Arch Virol. 2015 Oct;160(10):2629-32. doi: 10.1007/s00705-015-2547-3. Epub 2015 Aug 4. Arch Virol. 2015. PMID: 26234183
-
Genomic characterization of a turkey reovirus field strain by Next-Generation Sequencing.Infect Genet Evol. 2015 Jun;32:313-21. doi: 10.1016/j.meegid.2015.03.029. Epub 2015 Apr 1. Infect Genet Evol. 2015. PMID: 25841748 Free PMC article.
-
Pathogenicity and genomic characterization of a novel avian orthoreovius variant isolated from a vaccinated broiler flock in China.Avian Pathol. 2019 Aug;48(4):334-342. doi: 10.1080/03079457.2019.1600656. Epub 2019 Apr 18. Avian Pathol. 2019. PMID: 30915860
Cited by
-
Choosing the most suitable NGS technology to combine with a standardized viral enrichment protocol for obtaining complete avian orthoreovirus genomes from metagenomic samples.Front Bioinform. 2025 Feb 4;5:1498921. doi: 10.3389/fbinf.2025.1498921. eCollection 2025. Front Bioinform. 2025. PMID: 39967836 Free PMC article.
-
Simultaneous detection and partial molecular characterization of five RNA viruses associated with enteric disease in chickens: chicken astrovirus, avian nephritis virus, infectious bronchitis virus, avian rotavirus a and avian orthoreovirus, via multiplex RT-qPCR.Front Vet Sci. 2025 Apr 24;12:1536420. doi: 10.3389/fvets.2025.1536420. eCollection 2025. Front Vet Sci. 2025. PMID: 40343369 Free PMC article.
-
Enteric Viruses in Turkeys: A Systematic Review and Comparative Data Analysis.Viruses. 2025 Jul 24;17(8):1037. doi: 10.3390/v17081037. Viruses. 2025. PMID: 40872752 Free PMC article.
-
Optimizing the Conditions for Whole-Genome Sequencing of Avian Reoviruses.Viruses. 2023 Sep 16;15(9):1938. doi: 10.3390/v15091938. Viruses. 2023. PMID: 37766345 Free PMC article.
-
Molecular characterization and lineage analysis of canine astrovirus strains from dogs with gastrointestinal disease in Ecuador based on ORF-2 gene analysis.Front Vet Sci. 2025 Feb 3;11:1505903. doi: 10.3389/fvets.2024.1505903. eCollection 2024. Front Vet Sci. 2025. PMID: 39963367 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

