Loss of NF1 in Melanoma Confers Sensitivity to SYK Kinase Inhibition
- PMID: 36409827
- PMCID: PMC9845987
- DOI: 10.1158/0008-5472.CAN-22-0883
Loss of NF1 in Melanoma Confers Sensitivity to SYK Kinase Inhibition
Abstract
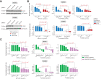
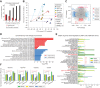
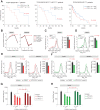
Neurofibromin 1 (NF1) loss of function (LoF) mutations are frequent in melanoma and drive hyperactivated RAS and tumor growth. NF1LoF melanoma cells, however, do not show consistent sensitivity to individual MEK, ERK, or PI3K/mTOR inhibitors. To identify more effective therapeutic strategies for treating NF1LoF melanoma, we performed a targeted kinase inhibitor screen. A tool compound named MTX-216 was highly effective in blocking NF1LoF melanoma growth in vitro and in vivo. Single-cell analysis indicated that drug-induced cytotoxicity was linked to effective cosuppression of proliferation marker Ki-67 and ribosomal protein S6 phosphorylation. The antitumor efficacy of MTX-216 was dependent on its ability to inhibit not only PI3K, its nominal target, but also SYK. MTX-216 suppressed expression of a group of genes that regulate mitochondrial electron transport chain and are associated with poor survival in patients with NF1LoF melanoma. Furthermore, combinations of inhibitors targeting either MEK or PI3K/mTOR with an independent SYK kinase inhibitor or SYK knockdown reduced the growth of NF1LoF melanoma cells. These studies provide a path to exploit SYK dependency to selectively target NF1LoF melanoma cells.
Significance: A kinase inhibitor screen identifies SYK as a targetable vulnerability in melanoma cells with NF1 loss of function.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures



Similar articles
-
Concurrent SOS1 and MEK suppression inhibits signaling and growth of NF1-null melanoma.Cell Rep Med. 2024 Nov 19;5(11):101818. doi: 10.1016/j.xcrm.2024.101818. Epub 2024 Nov 1. Cell Rep Med. 2024. PMID: 39488215 Free PMC article.
-
Mitogen-activated protein kinase dependency in BRAF/RAS wild-type melanoma: A rationale for combination inhibitors.Pigment Cell Melanoma Res. 2020 Mar;33(2):345-357. doi: 10.1111/pcmr.12824. Epub 2019 Sep 25. Pigment Cell Melanoma Res. 2020. PMID: 31518489
-
A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition.Cancer Discov. 2013 Mar;3(3):350-62. doi: 10.1158/2159-8290.CD-12-0470. Epub 2013 Jan 3. Cancer Discov. 2013. PMID: 23288408 Free PMC article.
-
Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):4015-20. doi: 10.1073/pnas.1216013110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431193 Free PMC article.
-
MEK inhibitors in oncology: a patent review (2015-Present).Expert Opin Ther Pat. 2017 Aug;27(8):887-906. doi: 10.1080/13543776.2017.1339688. Epub 2017 Jun 19. Expert Opin Ther Pat. 2017. PMID: 28594589 Review.
Cited by
-
Multivariate analysis of metabolic state vulnerabilities across diverse cancer contexts reveals synthetically lethal associations.bioRxiv [Preprint]. 2024 Jul 10:2023.11.28.569098. doi: 10.1101/2023.11.28.569098. bioRxiv. 2024. Update in: Cell Rep. 2024 Oct 22;43(10):114775. doi: 10.1016/j.celrep.2024.114775. PMID: 38076921 Free PMC article. Updated. Preprint.
-
Multivariate analysis of metabolic state vulnerabilities across diverse cancer contexts reveals synthetically lethal associations.Cell Rep. 2024 Oct 22;43(10):114775. doi: 10.1016/j.celrep.2024.114775. Epub 2024 Sep 20. Cell Rep. 2024. PMID: 39305483 Free PMC article.
-
Drug mechanism enrichment analysis improves prioritization of therapeutics for repurposing.BMC Bioinformatics. 2023 May 24;24(1):215. doi: 10.1186/s12859-023-05343-8. BMC Bioinformatics. 2023. PMID: 37226094 Free PMC article.
-
Cell Death Modalities in Therapy of Melanoma.Int J Mol Sci. 2025 Apr 8;26(8):3475. doi: 10.3390/ijms26083475. Int J Mol Sci. 2025. PMID: 40331942 Free PMC article. Review.
-
Advancing immunotherapy for melanoma: the critical role of single-cell analysis in identifying predictive biomarkers.Front Immunol. 2024 Jul 4;15:1435187. doi: 10.3389/fimmu.2024.1435187. eCollection 2024. Front Immunol. 2024. PMID: 39026661 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

