Target recognition by RNase E RNA-binding domain AR2 drives sRNA decay in the absence of PNPase
- PMID: 36409892
- PMCID: PMC9860253
- DOI: 10.1073/pnas.2208022119
Target recognition by RNase E RNA-binding domain AR2 drives sRNA decay in the absence of PNPase
Abstract
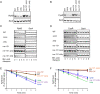
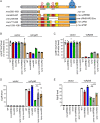
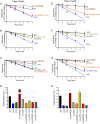
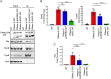
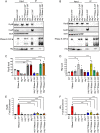
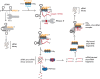
The C-terminal domain (CTD) of the major endoribonuclease RNase E not only serves as a scaffold for the central RNA decay machinery in gram-negative bacteria but also mediates coupled degradation of small regulatory RNAs (sRNAs) and their cognate target transcripts following RNA chaperone Hfq-facilitated sRNA-mRNA base pairing. Despite the crucial role of RNase E CTD in sRNA-dependent gene regulation, the contribution of particular residues within this domain in recruiting sRNAs and mRNAs upon base pairing remains unknown. We have previously shown that in Escherichia coli, the highly conserved 3'-5'-exoribonuclease polynucleotide phosphorylase (PNPase) paradoxically stabilizes sRNAs by limiting access of RNase E to Hfq-bound sRNAs and by degrading target mRNA fragments that would otherwise promote sRNA decay. Here, we report that in the absence of PNPase, the RNA-binding region AR2 in the CTD is required for RNase E to initiate degradation of the Hfq-dependent sRNAs CyaR and RyhB. Additionally, we show that introducing mutations in either hfq that disrupts target mRNA binding to Hfq or the AR2 coding region of rne impairs RNase E binding to sRNAs. Altogether, our data support a model where sRNAs are recruited via bound mRNA targets to RNase E by its AR2 domain after Hfq catalyzes sRNA-mRNA pairing. These results also support our conclusion that in a PNPase-deficient strain, more rapid decay of sRNAs occurs due to accelerated pairing with mRNA targets as a consequence of their accumulation. Our findings provide insights into the mechanisms by which sRNAs and mRNAs are regulated by RNase E.
Keywords: Hfq; RNase E; polynucleotide phosphorylase; sRNA; small RNA.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Wagner E. G. H., Romby P., Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015). - PubMed
-
- Holmqvist E., Berggren S., Rizvanovic A., RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. acta. Gene Regul. Mech. 1863, 194596 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

