Regulation of the DEAH/RHA helicase Prp43 by the G-patch factor Pfa1
- PMID: 36409901
- PMCID: PMC9860317
- DOI: 10.1073/pnas.2203567119
Regulation of the DEAH/RHA helicase Prp43 by the G-patch factor Pfa1
Abstract
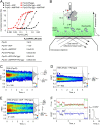
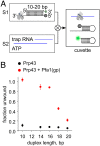
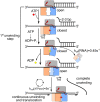
The DEAH/RHA helicase Prp43 remodels protein-RNA complexes during pre-messenger RNA (mRNA) splicing and ribosome biogenesis. The helicase activity and ATP turnover are intrinsically low and become activated by G-patch (gp) factors in the specific cellular context. The gp motif connects the helicase core to the flexible C-terminal domains, but it is unclear how this affects RecA domain movement during catalysis and the unwinding of RNA substrates. We developed single-molecule Förster Resonance Energy Transfer (smFRET) reporters to study RecA domain movements within Prp43 in real time. Without Pfa1(gp), the domains approach each other adopting predominantly a closed conformation. The addition of Pfa1(gp) induces an open state, which becomes even more prevalent during interaction with RNA. In the open state, Prp43 has reduced contacts with bound nucleotide and shows rapid adenosine diphosphate (ADP) release accelerating the transition from the weak (ADP) to the strong (apo) RNA binding state. Using smFRET labels on the RNA to probe substrate binding and unwinding, we demonstrate that Pfa1(gp) enables Prp43(ADP) to switch between RNA-bound and RNA-unbound states instead of dissociating from the RNA. ATP binding to the apo-enzyme induces the translocation along the RNA, generating the unwinding force required to melt proximal RNA structures. During ATP turnover, Pfa1(gp) stimulates alternating of the RecA domains between open and closed states. Consequently, the translocation becomes faster than dissociation from the substrate in the ADP state, allowing processive movement along the RNA. We provide a mechanistic model of DEAH/RHA helicase motility and reveal the principles of Prp43 regulation by G-patch proteins.
Keywords: DEAH/RHA helicases; G-patch proteins; molecular machines; ribosome biogenesis; splicing.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Conformational dynamics of the RNA binding channel regulates loading and translocation of the DEAH-box helicase Prp43.Nucleic Acids Res. 2023 Jul 7;51(12):6430-6442. doi: 10.1093/nar/gkad362. Nucleic Acids Res. 2023. PMID: 37167006 Free PMC article.
-
The G-patch activators Pfa1 and PINX1 exhibit different modes of interaction with the Prp43 RNA helicase.RNA Biol. 2021 Apr;18(4):510-522. doi: 10.1080/15476286.2020.1818458. Epub 2020 Oct 30. RNA Biol. 2021. PMID: 32882145 Free PMC article.
-
Structural basis for DEAH-helicase activation by G-patch proteins.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7159-7170. doi: 10.1073/pnas.1913880117. Epub 2020 Mar 16. Proc Natl Acad Sci U S A. 2020. PMID: 32179686 Free PMC article.
-
Regulation of DEAH/RHA helicases by G-patch proteins.Biomed Res Int. 2015;2015:931857. doi: 10.1155/2015/931857. Epub 2015 Jan 27. Biomed Res Int. 2015. PMID: 25692149 Free PMC article. Review.
-
Regulation of DEAH-box RNA helicases by G-patch proteins.Biol Chem. 2021 Jan 6;402(5):561-579. doi: 10.1515/hsz-2020-0338. Print 2021 Apr 27. Biol Chem. 2021. PMID: 33857358 Review.
Cited by
-
Dysfunction of Gpl1-Gih35-Wdr83 Complex in S. pombe Affects the Splicing of DNA Damage Repair Factors Resulting in Increased Sensitivity to DNA Damage.Int J Mol Sci. 2024 Apr 10;25(8):4192. doi: 10.3390/ijms25084192. Int J Mol Sci. 2024. PMID: 38673778 Free PMC article.
-
In vitro characterization of the yeast DEAH/RHA RNA helicase Dhr1.J Biol Chem. 2025 Apr;301(4):108366. doi: 10.1016/j.jbc.2025.108366. Epub 2025 Feb 28. J Biol Chem. 2025. PMID: 40024476 Free PMC article.
-
GPATCH4 regulates rRNA and snRNA 2'-O-methylation in both DHX15-dependent and DHX15-independent manners.Nucleic Acids Res. 2024 Feb 28;52(4):1953-1974. doi: 10.1093/nar/gkad1202. Nucleic Acids Res. 2024. PMID: 38113271 Free PMC article.
-
Conformational dynamics of the RNA binding channel regulates loading and translocation of the DEAH-box helicase Prp43.Nucleic Acids Res. 2023 Jul 7;51(12):6430-6442. doi: 10.1093/nar/gkad362. Nucleic Acids Res. 2023. PMID: 37167006 Free PMC article.
-
Cellular functions of eukaryotic RNA helicases and their links to human diseases.Nat Rev Mol Cell Biol. 2023 Oct;24(10):749-769. doi: 10.1038/s41580-023-00628-5. Epub 2023 Jul 20. Nat Rev Mol Cell Biol. 2023. PMID: 37474727 Review.
References
-
- Pyle A. M., Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 (2008). - PubMed
-
- Caruthers J. M., McKay D. B., Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 (2002). - PubMed
-
- Cordin O., Banroques J., Tanner N. K., Linder P., The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

