Escherichia coli O157:H7 senses microbiota-produced riboflavin to increase its virulence in the gut
- PMID: 36409903
- PMCID: PMC9860305
- DOI: 10.1073/pnas.2212436119
Escherichia coli O157:H7 senses microbiota-produced riboflavin to increase its virulence in the gut
Abstract
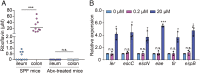
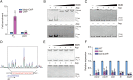
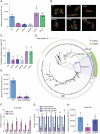
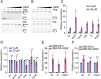
Riboflavin is produced by most commensal bacteria in the human colon, where enterohemorrhagic Escherichia coli (EHEC) colonizes and causes diseases. Sensing environmental signals to site-specifically express the type-III secretion system (T3SS), which injects effectors into host cells leading to intestinal colonization and disease, is key to the pathogenesis of EHEC. Here, we reveal that EHEC O157:H7, a dominant EHEC serotype frequently associated with severe diseases, acquired a previously uncharacterized two-component regulatory system rbfSR, which senses microbiota-produced riboflavin to directly activate the expression of LEE genes encoding the T3SS in the colon. rbfSR is present in O157:H7 and O145:H28 but absent from other EHEC serotypes. The binding site of RbfR through which it regulates LEE gene expression was identified and is conserved in all EHEC serotypes and Citrobacter rodentium, a surrogate for EHEC in mice. Introducing rbfSR into C. rodentium enabled bacteria to sense microbiota-produced riboflavin in the mouse colon to increase the expression of LEE genes, causing increased disease severity in mice. Phylogenic analysis showed that the O55:H7 ancestor of O157:H7 obtained rbfSR which has been kept in O157:H7 since then. Thus, acquiring rbfSR represents an essential step in the evolution of the highly pathogenic O157:H7. The expression of LEE genes and cell attachment ability of other EHEC serotypes in the presence of riboflavin significantly increased when rbfSR was introduced into them, indicating that those serotypes are ready to use RbfSR to increase their pathogenicity. This may present a potential public health issue as horizontal gene transfer is frequent in enteric bacteria.
Keywords: enterohaemorrhagic Escherichia coli; evolution; gut microbiota; two-component regulatory system; virulence regulation.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Kaper J. B., Nataro J. P., Mobley H. L., Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004). - PubMed
-
- Vallance B. A., Chan C., Robertson M. L., Finlay B. B., Enteropathogenic and enterohemorrhagic Escherichia coli infections: Emerging themes in pathogenesis and prevention. Can. J. Gastroenterol. 16, 771–778 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

