T cell and B cell antigen receptors share a conserved core transmembrane structure
- PMID: 36409917
- PMCID: PMC9860311
- DOI: 10.1073/pnas.2208058119
T cell and B cell antigen receptors share a conserved core transmembrane structure
Abstract
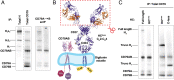
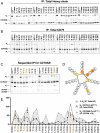
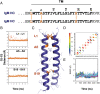
The B cell and T cell antigen receptors (BCR and TCR) share a common architecture in which variable dimeric antigen-binding modules assemble with invariant dimeric signaling modules to form functional receptor complexes. In the TCR, a highly conserved T cell receptor αβ (TCRαβ) transmembrane (TM) interface forms a rigid structure around which its three dimeric signaling modules assemble through well-characterized polar interactions. Noting that the key features stabilizing this TCRαβ TM interface also appear with high evolutionary conservation in the TM sequences of the membrane immunoglobulin (mIg) heavy chains that form the BCR's homodimeric antigen-binding module, we asked whether the BCR contained an analogous TM structure. Using an unbiased biochemical and computational modeling approach, we found that the mouse IgM BCR forms a core TM structure that is remarkably similar to that of the TCR. This structure is reinforced by a network of interhelical hydrogen bonds, and our model is nearly identical to the arrangement observed in the just-released cryo-electron microscopy (cryo-EM) structures of intact human BCRs. Our biochemical analysis shows that the integrity of this TM structure is vital for stable assembly with the BCR signaling module CD79AB in the B cell endoplasmic reticulum, and molecular dynamics simulations indicate that BCRs of all five isotypes can form comparable structures. These results demonstrate that, despite their many differences in composition, complexity, and ligand type, TCRs and BCRs rely on a common core TM structure that has been shaped by evolution for optimal receptor assembly and stability in the cell membrane.
Keywords: B cell receptor; antigen receptor; receptor assembly; receptor structure; transmembrane.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Structural Conservation and Effects of Alterations in T Cell Receptor Transmembrane Interfaces.Biophys J. 2018 Mar 13;114(5):1030-1035. doi: 10.1016/j.bpj.2018.01.004. Epub 2018 Jan 29. Biophys J. 2018. PMID: 29395047 Free PMC article.
-
A conserved αβ transmembrane interface forms the core of a compact T-cell receptor-CD3 structure within the membrane.Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):E6649-E6658. doi: 10.1073/pnas.1611445113. Epub 2016 Oct 10. Proc Natl Acad Sci U S A. 2016. PMID: 27791034 Free PMC article.
-
Structural Model of the mIgM B-Cell Receptor Transmembrane Domain From Self-Association Molecular Dynamics Simulations.Front Immunol. 2018 Dec 17;9:2947. doi: 10.3389/fimmu.2018.02947. eCollection 2018. Front Immunol. 2018. PMID: 30619307 Free PMC article.
-
Composition and function of T-cell receptor and B-cell receptor complexes on precursor lymphocytes.Curr Opin Immunol. 1996 Apr;8(2):181-90. doi: 10.1016/s0952-7915(96)80056-2. Curr Opin Immunol. 1996. PMID: 8725941 Review.
-
Molecular mechanisms for the assembly of the T cell receptor-CD3 complex.Mol Immunol. 2004 Apr;40(18):1295-305. doi: 10.1016/j.molimm.2003.11.017. Mol Immunol. 2004. PMID: 15072848 Free PMC article. Review.
Cited by
-
Deep mutational scanning reveals transmembrane features governing surface expression of the B cell antigen receptor.Front Immunol. 2024 Jul 23;15:1426795. doi: 10.3389/fimmu.2024.1426795. eCollection 2024. Front Immunol. 2024. PMID: 39108267 Free PMC article.
-
Combination of High-Resolution Structures for the B Cell Receptor and Co-Receptors Provides an Understanding of Their Interactions with Therapeutic Antibodies.Cancers (Basel). 2023 May 23;15(11):2881. doi: 10.3390/cancers15112881. Cancers (Basel). 2023. PMID: 37296844 Free PMC article. Review.
-
BCR, not TCR, repertoire diversity is associated with favorable COVID-19 prognosis.Front Immunol. 2024 Oct 28;15:1405013. doi: 10.3389/fimmu.2024.1405013. eCollection 2024. Front Immunol. 2024. PMID: 39530088 Free PMC article.
References
-
- Berry R., Call M. E., Modular activating receptors in innate and adaptive immunity. Biochemistry 56, 1383–1402 (2017). - PubMed
-
- Reth M., Antigen receptor tail clue. Nature 338, 383 (1989). - PubMed
-
- Alt F. W., et al. , Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3’ ends. Cell 20, 293–301 (1980). - PubMed
-
- Early P., et al. , Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20, 313–319 (1980). - PubMed
-
- Hombach J., Tsubata T., Leclercq L., Stappert H., Reth M., Molecular components of the B-cell antigen receptor complex of the IgM class. Nature 343, 760–762 (1990). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

