Dysregulated Amino Acid Sensing Drives Colorectal Cancer Growth and Metabolic Reprogramming Leading to Chemoresistance
- PMID: 36410445
- PMCID: PMC10448739
- DOI: 10.1053/j.gastro.2022.11.014
Dysregulated Amino Acid Sensing Drives Colorectal Cancer Growth and Metabolic Reprogramming Leading to Chemoresistance
Abstract
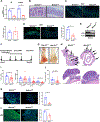
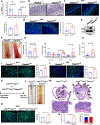
Background & aims: Colorectal cancer (CRC) is a devastating disease that is highly modulated by dietary nutrients. Mechanistic target of rapamycin complex 1 (mTORC1) contributes to tumor growth and limits therapy responses. Growth factor signaling is a major mechanism of mTORC1 activation. However, compensatory pathways exist to sustain mTORC1 activity after therapies that target oncogenic growth factor signaling. Amino acids potently activate mTORC1 via amino acid-sensing GTPase activity towards Rags (GATOR). The role of amino acid-sensing pathways in CRC is unclear.
Methods: Human colon cancer cell lines, preclinical intestinal epithelial-specific GATOR1 and GATOR2 knockout mice subjected to colitis-induced or sporadic colon tumor models, small interfering RNA screening targeting regulators of mTORC1, and tissues of patients with CRC were used to assess the role of amino acid sensing in CRC.
Results: We identified loss-of-function mutations of the GATOR1 complex in CRC and showed that altered expression of amino acid-sensing pathways predicted poor patient outcomes. We showed that dysregulated amino acid-sensing induced mTORC1 activation drives colon tumorigenesis in multiple mouse models. We found amino acid-sensing pathways to be essential in the cellular reprogramming of chemoresistance, and chemotherapeutic-resistant patients with colon cancer exhibited de-regulated amino acid sensing. Limiting amino acids in in vitro and in vivo models (low-protein diet) reverted drug resistance, revealing a metabolic vulnerability.
Conclusions: Our findings suggest a critical role for amino acid-sensing pathways in driving CRC and highlight the translational implications of dietary protein intervention in CRC.
Keywords: 5-Fluorouracil; Depdc5; Sestrin 2; Wdr24; mTORC1.
Copyright © 2023 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

