Multi-omics signatures of the human early life exposome
- PMID: 36411288
- PMCID: PMC9678903
- DOI: 10.1038/s41467-022-34422-2
Multi-omics signatures of the human early life exposome
Abstract
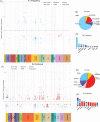
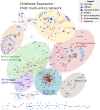
Environmental exposures during early life play a critical role in life-course health, yet the molecular phenotypes underlying environmental effects on health are poorly understood. In the Human Early Life Exposome (HELIX) project, a multi-centre cohort of 1301 mother-child pairs, we associate individual exposomes consisting of >100 chemical, outdoor, social and lifestyle exposures assessed in pregnancy and childhood, with multi-omics profiles (methylome, transcriptome, proteins and metabolites) in childhood. We identify 1170 associations, 249 in pregnancy and 921 in childhood, which reveal potential biological responses and sources of exposure. Pregnancy exposures, including maternal smoking, cadmium and molybdenum, are predominantly associated with child DNA methylation changes. In contrast, childhood exposures are associated with features across all omics layers, most frequently the serum metabolome, revealing signatures for diet, toxic chemical compounds, essential trace elements, and weather conditions, among others. Our comprehensive and unique resource of all associations ( https://helixomics.isglobal.org/ ) will serve to guide future investigation into the biological imprints of the early life exposome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

