A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings
- PMID: 36411354
- PMCID: PMC9712112
- DOI: 10.1038/s41564-022-01263-0
A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings
Abstract
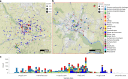
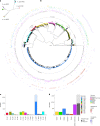
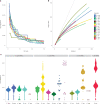
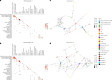
The Klebsiella group, found in humans, livestock, plants, soil, water and wild animals, is genetically and ecologically diverse. Many species are opportunistic pathogens and can harbour diverse classes of antimicrobial resistance genes. Healthcare-associated Klebsiella pneumoniae clones that are non-susceptible to carbapenems can spread rapidly, representing a high public health burden. Here we report an analysis of 3,482 genome sequences representing 15 Klebsiella species sampled over a 17-month period from a wide range of clinical, community, animal and environmental settings in and around the Italian city of Pavia. Northern Italy is a hotspot for hospital-acquired carbapenem non-susceptible Klebsiella and thus a pertinent setting to examine the overlap between isolates in clinical and non-clinical settings. We found no genotypic or phenotypic evidence for non-susceptibility to carbapenems outside the clinical environment. Although we noted occasional transmission between clinical and non-clinical settings, our data point to a limited role of animal and environmental reservoirs in the human acquisition of Klebsiella spp. We also provide a detailed genus-wide view of genomic diversity and population structure, including the identification of new groups.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures