Mucosal-associated invariant T cells restrict reactive oxidative damage and preserve meningeal barrier integrity and cognitive function
- PMID: 36411380
- PMCID: PMC10202031
- DOI: 10.1038/s41590-022-01349-1
Mucosal-associated invariant T cells restrict reactive oxidative damage and preserve meningeal barrier integrity and cognitive function
Abstract
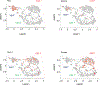
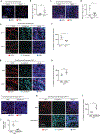
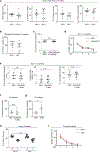
Increasing evidence indicates close interaction between immune cells and the brain, revising the traditional view of the immune privilege of the brain. However, the specific mechanisms by which immune cells promote normal neural function are not entirely understood. Mucosal-associated invariant T cells (MAIT cells) are a unique type of innate-like T cell with molecular and functional properties that remain to be better characterized. In the present study, we report that MAIT cells are present in the meninges and express high levels of antioxidant molecules. MAIT cell deficiency in mice results in the accumulation of reactive oxidative species in the meninges, leading to reduced expression of junctional protein and meningeal barrier leakage. The presence of MAIT cells restricts neuroinflammation in the brain and preserves learning and memory. Together, our work reveals a new functional role for MAIT cells in the meninges and suggests that meningeal immune cells can help maintain normal neural function by preserving meningeal barrier homeostasis and integrity.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures

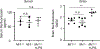








Comment in
-
Meningeal MAIT cells preserve brain homeostasis.Nat Rev Immunol. 2023 Jan;23(1):4. doi: 10.1038/s41577-022-00824-y. Nat Rev Immunol. 2023. PMID: 36477414 No abstract available.
-
Meningeal MAIT cells maintain meningeal and brain function.Nat Immunol. 2022 Dec;23(12):1659-1661. doi: 10.1038/s41590-022-01368-y. Nat Immunol. 2022. PMID: 36494576 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

