Molecular and spatial heterogeneity of microglia in Rasmussen encephalitis
- PMID: 36411471
- PMCID: PMC9677917
- DOI: 10.1186/s40478-022-01472-y
Molecular and spatial heterogeneity of microglia in Rasmussen encephalitis
Abstract
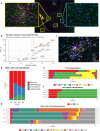
Rasmussen encephalitis (RE) is a rare childhood neurological disease characterized by progressive unilateral loss of function, hemispheric atrophy and drug-resistant epilepsy. Affected brain tissue shows signs of infiltrating cytotoxic T-cells, microglial activation, and neuronal death, implicating an inflammatory disease process. Recent studies have identified molecular correlates of inflammation in RE, but cell-type-specific mechanisms remain unclear. We used single-nucleus RNA-sequencing (snRNA-seq) to assess gene expression across multiple cell types in brain tissue resected from two children with RE. We found transcriptionally distinct microglial populations enriched in RE compared to two age-matched individuals with unaffected brain tissue and two individuals with Type I focal cortical dysplasia (FCD). Specifically, microglia in RE tissues demonstrated increased expression of genes associated with cytokine signaling, interferon-mediated pathways, and T-cell activation. We extended these findings using spatial proteomic analysis of tissue from four surgical resections to examine expression profiles of microglia within their pathological context. Microglia that were spatially aggregated into nodules had increased expression of dynamic immune regulatory markers (PD-L1, CD14, CD11c), T-cell activation markers (CD40, CD80) and were physically located near distinct CD4+ and CD8+ lymphocyte populations. These findings help elucidate the complex immune microenvironment of RE.
Keywords: Cytokine signaling; Encephalitis; Epilepsy; Inflammation; Microglial nodules; Single nucleus RNA-seq; Spatial proteomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

