Amplicon-based assessment of bacterial diversity and community structure in three tropical forest soils in Kenya
- PMID: 36411924
- PMCID: PMC9674510
- DOI: 10.1016/j.heliyon.2022.e11577
Amplicon-based assessment of bacterial diversity and community structure in three tropical forest soils in Kenya
Abstract
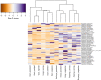
Forest soils provide a multitude of habitats for diverse communities of bacteria. In this study, we selected three tropical forests in Kenya to determine the diversity and community structure of soil bacteria inhabiting these regions. Kakamega and Irangi are rainforests, whereas Gazi Bay harbors mangrove forests. The three natural forests occupy different altitudinal zones and differ in their environmental characteristics. Soil samples were collected from a total of 12 sites and soil physicochemical parameters for each sampling site were analyzed. We used an amplicon-based Illumina high-throughput sequencing approach. Total community DNA was extracted from individual samples using the phenol-chloroform method. The 16S ribosomal RNA gene segment spanning the V4 region was amplified using the Illumina MiSeq platform. Diversity indices, rarefaction curves, hierarchical clustering, principal component analysis (PCA), and non-metric multidimensional scaling (NMDS) analyses were performed in R software. A total of 13,410 OTUs were observed at 97% sequence similarity. Bacterial communities were dominated by Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Acidobacteria in both rainforest and mangrove sampling sites. Alpha diversity indices and species richness were higher in Kakamega and Irangi rainforests compared to mangroves in Gazi Bay. The composition of bacterial communities within and between the three forests was also significantly differentiated (R = 0.559, p = 0.007). Clustering in both PCA and NMDS plots showed that each sampling site had a distinct bacterial community profile. The NMDS analysis also indicated that soil EC, sodium, sulfur, magnesium, boron, and manganese contributed significantly to the observed variation in the bacterial community structure. Overall, this study demonstrated the presence of diverse taxa and heterogeneous community structures of soil bacteria inhabiting three tropical forests of Kenya. Our results also indicated that variation in soil chemical parameters was the major driver of the observed bacterial diversity and community structure in these forests.
Keywords: 16S rRNA; Community structure; Diversity; Soil bacteria; Soil chemical parameters; Tropical forests.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Aislabie J., Deslippe J.R., Dymond J. Soil microbes and their contribution to soil services. Ecosyst. Serv. New Zealand–Cond. Trends. 2013;1(12):143–161. Manaaki Whenua Press, Lincoln, New Zealand.
-
- Amoo A.E., Babalola O.O. Impact of land use on bacterial diversity and community structure in temperate pine and indigenous forest soils. Diversity. 2019;11(11):217.
-
- Bañeras Vives L., Ruiz Rueda O., López i Flores R., Quintana Pou X., Hallin S. The role of plant type and salinity in the selection for the denitrifying community structure in the rhizosphere of wetland vegetation. Int. Microbiol. 2012;15(2):89–99. - PubMed
LinkOut - more resources
Full Text Sources

