Age acquired skewed X chromosome inactivation is associated with adverse health outcomes in humans
- PMID: 36412098
- PMCID: PMC9681199
- DOI: 10.7554/eLife.78263
Age acquired skewed X chromosome inactivation is associated with adverse health outcomes in humans
Abstract
Background: Ageing is a heterogenous process characterised by cellular and molecular hallmarks, including changes to haematopoietic stem cells and is a primary risk factor for chronic diseases. X chromosome inactivation (XCI) randomly transcriptionally silences either the maternal or paternal X in each cell of 46, XX females to balance the gene expression with 46, XY males. Age acquired XCI-skew describes the preferential selection of cells across a tissue resulting in an imbalance of XCI, which is particularly prevalent in blood tissues of ageing females, and yet its clinical consequences are unknown.
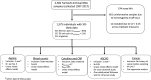
Methods: We assayed XCI in 1575 females from the TwinsUK population cohort using DNA extracted from whole blood. We employed prospective, cross-sectional, and intra-twin study designs to characterise the relationship of XCI-skew with molecular and cellular measures of ageing, cardiovascular disease risk, and cancer diagnosis.
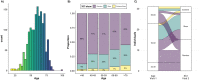
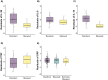
Results: We demonstrate that XCI-skew is independent of traditional markers of biological ageing and is associated with a haematopoietic bias towards the myeloid lineage. Using an atherosclerotic cardiovascular disease risk score, which captures traditional risk factors, XCI-skew is associated with an increased cardiovascular disease risk both cross-sectionally and within XCI-skew discordant twin pairs. In a prospective 10 year follow-up study, XCI-skew is predictive of future cancer incidence.
Conclusions: Our study demonstrates that age acquired XCI-skew captures changes to the haematopoietic stem cell population and has clinical potential as a unique biomarker of chronic disease risk.
Funding: KSS acknowledges funding from the Medical Research Council [MR/M004422/1 and MR/R023131/1]. JTB acknowledges funding from the ESRC [ES/N000404/1]. MM acknowledges funding from the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London.
Keywords: X chromosome inactivation; cancer; cardiovascular disease; epidemiology; genetics; genomics; global health; haematopoiesis; human.
© 2022, Roberts et al.
Conflict of interest statement
AR, AM, AA, AZ, JE, MT, RB, XZ, CC, RC, MM, JB, CW, TV, KS No competing interests declared, CS Consultant for Zoe Ltd
Figures


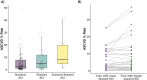
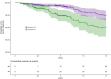
Similar articles
-
Genome-wide association study meta-analysis of dizygotic twinning illuminates genetic regulation of female fecundity.Hum Reprod. 2024 Jan 5;39(1):240-257. doi: 10.1093/humrep/dead247. Hum Reprod. 2024. PMID: 38052102 Free PMC article.
-
Haematopoietic stem cell-derived immune cells have reduced X chromosome inactivation skewing in systemic lupus erythematosus.Ann Rheum Dis. 2024 Sep 30;83(10):1315-1321. doi: 10.1136/ard-2024-225585. Ann Rheum Dis. 2024. PMID: 38937070 Free PMC article.
-
X-chromosome inactivation patterns depend on age and tissue but not conception method in humans.Chromosome Res. 2023 Jan 25;31(1):4. doi: 10.1007/s10577-023-09717-9. Chromosome Res. 2023. PMID: 36695960 Free PMC article.
-
X chromosome inactivation in human pluripotent stem cells as a model for human development: back to the drawing board?Hum Reprod Update. 2017 Sep 1;23(5):520-532. doi: 10.1093/humupd/dmx015. Hum Reprod Update. 2017. PMID: 28582519 Review.
-
Sex differences in susceptibility to substance use disorder: Role for X chromosome inactivation and escape?Mol Cell Neurosci. 2023 Jun;125:103859. doi: 10.1016/j.mcn.2023.103859. Epub 2023 May 18. Mol Cell Neurosci. 2023. PMID: 37207894 Free PMC article. Review.
Cited by
-
Escape of Kdm6a from X chromosome is detrimental to ischemic brains via IRF5 signaling.Res Sq [Preprint]. 2024 Sep 27:rs.3.rs-4986866. doi: 10.21203/rs.3.rs-4986866/v1. Res Sq. 2024. Update in: Transl Stroke Res. 2025 Jan 3. doi: 10.1007/s12975-024-01321-1. PMID: 39399684 Free PMC article. Updated. Preprint.
-
Population analyses of mosaic X chromosome loss identify genetic drivers and widespread signatures of cellular selection.medRxiv [Preprint]. 2023 Jan 31:2023.01.28.23285140. doi: 10.1101/2023.01.28.23285140. medRxiv. 2023. PMID: 36778285 Free PMC article. Preprint.
-
A whole-organism landscape of X-inactivation in humans.Elife. 2025 Jul 17;14:RP102701. doi: 10.7554/eLife.102701. Elife. 2025. PMID: 40673498 Free PMC article.
-
Establishing the Link between X-Chromosome Aberrations and TP53 Status, with Breast Cancer Patient Outcomes.Cells. 2023 Sep 11;12(18):2245. doi: 10.3390/cells12182245. Cells. 2023. PMID: 37759468 Free PMC article.
-
DNA methylation differences between the female and male X chromosomes in human brain.bioRxiv [Preprint]. 2024 Apr 17:2024.04.16.589778. doi: 10.1101/2024.04.16.589778. bioRxiv. 2024. PMID: 38659923 Free PMC article. Preprint.
References
-
- Arbel Y, Finkelstein A, Halkin A, Birati EY, Revivo M, Zuzut M, Shevach A, Berliner S, Herz I, Keren G, Banai S. Neutrophil/lymphocyte ratio is related to the severity of coronary artery disease and clinical outcome in patients undergoing angiography. Atherosclerosis. 2012;225:456–460. doi: 10.1016/j.atherosclerosis.2012.09.009. - DOI - PubMed
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. Journal of Statistical Software. 2015;67:jss.v067.i01. doi: 10.18637/jss.v067.i01. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

