Navigating the Multiverse of Antisense RNAs: The Transcription- and RNA-Dependent Dimension
- PMID: 36412909
- PMCID: PMC9680235
- DOI: 10.3390/ncrna8060074
Navigating the Multiverse of Antisense RNAs: The Transcription- and RNA-Dependent Dimension
Abstract
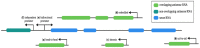
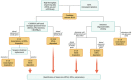
Evidence accumulated over the past decades shows that the number of identified antisense transcripts is continuously increasing, promoting them from transcriptional noise to real genes with specific functions. Indeed, recent studies have begun to unravel the complexity of the antisense RNA (asRNA) world, starting from the multidimensional mechanisms that they can exert in physiological and pathological conditions. In this review, we discuss the multiverse of the molecular functions of asRNAs, describing their action through transcription-dependent and RNA-dependent mechanisms. Then, we report the workflow and methodologies to study and functionally characterize single asRNA candidates.
Keywords: RNA-dependent mechanism; antisense RNA (asRNA); functional study; long non-coding RNA (lncRNA); transcription-dependent mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Antisense transcription and its roles in adaption to environmental stress in E. coli.bioRxiv [Preprint]. 2023 Mar 24:2023.03.23.533988. doi: 10.1101/2023.03.23.533988. bioRxiv. 2023. PMID: 36993172 Free PMC article. Preprint.
-
Antisense transcription is pervasive but rarely conserved in enteric bacteria.mBio. 2012 Aug 7;3(4):e00156-12. doi: 10.1128/mBio.00156-12. Print 2012. mBio. 2012. PMID: 22872780 Free PMC article.
-
[Non-coding Natural Antisense RNA: Mechanisms of Action in the Regulation of Target Gene Expression and Its Clinical Implications].Yakugaku Zasshi. 2020;140(5):687-700. doi: 10.1248/yakushi.20-00002. Yakugaku Zasshi. 2020. PMID: 32378673 Review. Japanese.
-
Bacterial antisense RNAs are mainly the product of transcriptional noise.Sci Adv. 2016 Mar 4;2(3):e1501363. doi: 10.1126/sciadv.1501363. eCollection 2016 Mar. Sci Adv. 2016. PMID: 26973873 Free PMC article.
-
Modulation of Bacterial Fitness and Virulence Through Antisense RNAs.Front Cell Infect Microbiol. 2021 Feb 11;10:596277. doi: 10.3389/fcimb.2020.596277. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33747974 Free PMC article. Review.
Cited by
-
KDM7A-DT induces genotoxic stress, tumorigenesis, and progression of p53 missense mutation-associated invasive breast cancer.Front Oncol. 2024 May 2;14:1227151. doi: 10.3389/fonc.2024.1227151. eCollection 2024. Front Oncol. 2024. PMID: 38756663 Free PMC article.
-
Integrating datasets to dissect NFYC-AS1 RNA- and transcription-dependent functions: comparative transcriptome profiling of knockdown strategies.Data Brief. 2025 Apr 15;60:111565. doi: 10.1016/j.dib.2025.111565. eCollection 2025 Jun. Data Brief. 2025. PMID: 40322504 Free PMC article.
-
Targeting lncRNAs of colorectal cancers with natural products.Front Pharmacol. 2023 Jan 9;13:1050032. doi: 10.3389/fphar.2022.1050032. eCollection 2022. Front Pharmacol. 2023. PMID: 36699052 Free PMC article. Review.
-
An antisense-long-noncoding-RNA modulates p75NTR expression levels during neuronal polarization.iScience. 2024 Dec 10;28(1):111566. doi: 10.1016/j.isci.2024.111566. eCollection 2025 Jan 17. iScience. 2024. PMID: 39811648 Free PMC article.
-
Study of lncRNAs in Pediatric Neurological Diseases: Methods, Analysis of the State-of-Art and Possible Therapeutic Implications.Pharmaceuticals (Basel). 2023 Nov 16;16(11):1616. doi: 10.3390/ph16111616. Pharmaceuticals (Basel). 2023. PMID: 38004481 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

