Machine learning approaches for electronic health records phenotyping: a methodical review
- PMID: 36413056
- PMCID: PMC9846699
- DOI: 10.1093/jamia/ocac216
Machine learning approaches for electronic health records phenotyping: a methodical review
Abstract
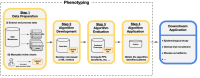
Objective: Accurate and rapid phenotyping is a prerequisite to leveraging electronic health records for biomedical research. While early phenotyping relied on rule-based algorithms curated by experts, machine learning (ML) approaches have emerged as an alternative to improve scalability across phenotypes and healthcare settings. This study evaluates ML-based phenotyping with respect to (1) the data sources used, (2) the phenotypes considered, (3) the methods applied, and (4) the reporting and evaluation methods used.
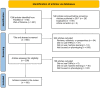
Materials and methods: We searched PubMed and Web of Science for articles published between 2018 and 2022. After screening 850 articles, we recorded 37 variables on 100 studies.
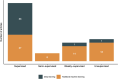
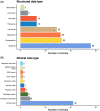
Results: Most studies utilized data from a single institution and included information in clinical notes. Although chronic conditions were most commonly considered, ML also enabled the characterization of nuanced phenotypes such as social determinants of health. Supervised deep learning was the most popular ML paradigm, while semi-supervised and weakly supervised learning were applied to expedite algorithm development and unsupervised learning to facilitate phenotype discovery. ML approaches did not uniformly outperform rule-based algorithms, but deep learning offered a marginal improvement over traditional ML for many conditions.
Discussion: Despite the progress in ML-based phenotyping, most articles focused on binary phenotypes and few articles evaluated external validity or used multi-institution data. Study settings were infrequently reported and analytic code was rarely released.
Conclusion: Continued research in ML-based phenotyping is warranted, with emphasis on characterizing nuanced phenotypes, establishing reporting and evaluation standards, and developing methods to accommodate misclassified phenotypes due to algorithm errors in downstream applications.
Keywords: cohort identification; electronic health records; machine learning; phenotyping.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Institute of Medicine, Roundtable on Value and Science-Driven Health Care. Clinical Data as the Basic Staple of Health Learning: Creating and Protecting a Public Good: Workshop Summary. Washington, DC: National Academies Press; 2011. - PubMed
-
- Li R, Chen Y, Ritchie MD, et al.Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet 2020; 21 (8): 493–502. - PubMed

