Feasibility and Potential of Transcriptomic Analysis Using the NanoString nCounter Technology to Aid the Classification of Rejection in Kidney Transplant Biopsies
- PMID: 36413151
- PMCID: PMC10065817
- DOI: 10.1097/TP.0000000000004372
Feasibility and Potential of Transcriptomic Analysis Using the NanoString nCounter Technology to Aid the Classification of Rejection in Kidney Transplant Biopsies
Abstract
Background: Transcriptome analysis could be an additional diagnostic parameter in diagnosing kidney transplant (KTx) rejection. Here, we assessed feasibility and potential of NanoString nCounter analysis of KTx biopsies to aid the classification of rejection in clinical practice using both the Banff-Human Organ Transplant (B-HOT) panel and a customized antibody-mediated rejection (AMR)-specific NanoString nCounter Elements (Elements) panel. Additionally, we explored the potential for the classification of KTx rejection building and testing a classifier within our dataset.
Methods: Ninety-six formalin-fixed paraffin-embedded KTx biopsies were retrieved from the archives of the ErasmusMC Rotterdam and the University Hospital Cologne. Biopsies with AMR, borderline or T cell-mediated rejections (BLorTCMR), and no rejection were compared using the B-HOT and Elements panels.
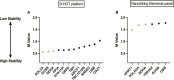
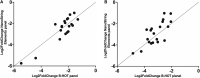
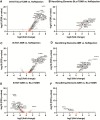
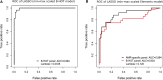
Results: High correlation between gene expression levels was found when comparing the 2 chemistries pairwise (r = 0.76-0.88). Differential gene expression (false discovery rate; P < 0.05) was identified in biopsies diagnosed with AMR (B-HOT: 294; Elements: 76) and BLorTCMR (B-HOT: 353; Elements: 57) compared with no rejection. Using the most predictive genes from the B-HOT analysis and the Element analysis, 2 least absolute shrinkage and selection operators-based regression models to classify biopsies as AMR versus no AMR (BLorTCMR or no rejection) were developed achieving an receiver-operating-characteristic curve of 0.994 and 0.894, sensitivity of 0.821 and 0.480, and specificity of 1.00 and 0.979, respectively, during cross-validation.
Conclusions: Transcriptomic analysis is feasible on KTx biopsies previously used for diagnostic purposes. The B-HOT panel has the potential to differentiate AMR from BLorTCMR or no rejection and could prove valuable in aiding kidney transplant rejection classification.
Copyright © 2022 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
M.C.C.v.G. has Astellas project funding paid to the Erasmus MC. J.U.B. is advisor for Sanofi. The other authors declare no conflicts of interest.
Figures
References
-
- Furness PN, Taub N, Assmann KJ, et al. . International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol. 2003;27:805–810. - PubMed
-
- Marcussen N, Olsen TS, Benediktsson H, et al. . Reproducibility of the Banff classification of renal allograft pathology. Inter- and intraobserver variation. Transplantation. 1995;60:1083–1089. - PubMed
-
- Haas M, Sis B, Racusen LC, et al. ; Banff meeting report writing committee. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant. 2014;14:272–283. - PubMed
-
- Adam B, Afzali B, Dominy KM, et al. . Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin Transplant. 2016;30:295–305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

