Intelectin-1 binds and alters the localization of the mucus barrier-modifying bacterium Akkermansia muciniphila
- PMID: 36413219
- PMCID: PMC9683900
- DOI: 10.1084/jem.20211938
Intelectin-1 binds and alters the localization of the mucus barrier-modifying bacterium Akkermansia muciniphila
Abstract
Intelectin-1 (ITLN1) is a lectin secreted by intestinal epithelial cells (IECs) and upregulated in human ulcerative colitis (UC). We investigated how ITLN1 production is regulated in IECs and the biological effects of ITLN1 at the host-microbiota interface using mouse models. Our data show that ITLN1 upregulation in IECs from UC patients is a consequence of activating the unfolded protein response. Analysis of microbes coated by ITLN1 in vivo revealed a restricted subset of microorganisms, including the mucolytic bacterium Akkermansia muciniphila. Mice overexpressing intestinal ITLN1 exhibited decreased inner colonic mucus layer thickness and closer apposition of A. muciniphila to the epithelial cell surface, similar to alterations reported in UC. The changes in the inner mucus layer were microbiota and A. muciniphila dependent and associated with enhanced sensitivity to chemically induced and T cell-mediated colitis. We conclude that by determining the localization of a select group of bacteria to the mucus layer, ITLN1 modifies this critical barrier. Together, these findings may explain the impact of ITLN1 dysregulation on UC pathogenesis.
© 2022 Matute et al.
Conflict of interest statement
Disclosures: G.M. Fuhler reported personal fees from Janssen and Galapagos during the conduct of the study. F. Tran reported personal fees from Abbvie, Janssen, and Falk outside the submitted work. J.R. White reported “other” from Resphera Biosciences LLC outside the submitted work. J.N. Glickman reported being an employee of PathAI, Inc. No other disclosures were reported.
Figures





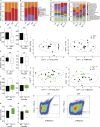

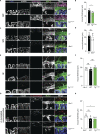
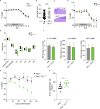

Comment in
-
Lectin recruits pathogenic bugs.J Exp Med. 2023 Jan 2;220(1):e20221732. doi: 10.1084/jem.20221732. Epub 2022 Nov 22. J Exp Med. 2023. PMID: 36413218 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R37 DK044319/DK/NIDDK NIH HHS/United States
- R01 DK088199/DK/NIDDK NIH HHS/United States
- R01 AI042347/AI/NIAID NIH HHS/United States
- R01 DK051362/DK/NIDDK NIH HHS/United States
- DK044319/NH/NIH HHS/United States
- MC_PC_MR/S025952/1/MRC_/Medical Research Council/United Kingdom
- R01 DE022586/DE/NIDCR NIH HHS/United States
- R01 DK068271/DK/NIDDK NIH HHS/United States
- MR/S036377/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/7/MRC_/Medical Research Council/United Kingdom
- R01 DK044319/DK/NIDDK NIH HHS/United States
- R56 DK053056/DK/NIDDK NIH HHS/United States
- 206194 /WT_/Wellcome Trust/United Kingdom
- R01 DK053056/DK/NIDDK NIH HHS/United States
- R01 DK061931/DK/NIDDK NIH HHS/United States
- 206194/WT_/Wellcome Trust/United Kingdom
- K12 HD000850/HD/NICHD NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- MC_UU_12010/7/MRC_/Medical Research Council/United Kingdom
- 106260/Z/14/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

