Unfolded protein response and angiogenesis in malignancies
- PMID: 36414127
- PMCID: PMC10167724
- DOI: 10.1016/j.bbcan.2022.188839
Unfolded protein response and angiogenesis in malignancies
Abstract
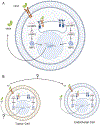
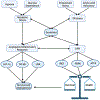
Cellular stress, arising from accumulation of unfolded proteins, occurs frequently in rapidly proliferating cancer cells. This cellular stress, in turn, activates the unfolded protein response (UPR), an interconnected set of signal transduction pathways that alleviate the proteostatic stress. The UPR is implicated in cancer cell survival and proliferation through upregulation of pro-tumorigenic pathways that ultimately promote malignant metabolism and neoangiogenesis. Here, we reviewed mechanisms of signaling crosstalk between the UPR and angiogenesis pathways, as well as transmissible ER stress and the role in tumor growth and development. To characterize differences in UPR and UPR-mediated angiogenesis in malignancy, we employed a data mining approach using patient tumor data from The Cancer Genome Atlas (TCGA). The analysis of TCGA revealed differences in UPR between malignant samples versus their non-malignant counterparts.
Keywords: ATF6; IRE1α; PERK; Tumor microenvironment; Unfolded protein response; XBP1.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Zeng L, Xiao Q, Chen M, Margariti A, Martin D, Ivetic A, Xu H, Mason J, Wang W, Cockerill G, Mori K, Li JY, Chien S, Hu Y, Xu Q, Vascular endothelial cell growth-activated XBP1 splicing in endothelial cells is crucial for angiogenesis, Circulation 127 (16) (2013) 1712–1722. - PubMed
-
- Chevet E, Hetz C, Samali A, Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis, Cancer Discov. 5 (6) (2015) 586–597. - PubMed
-
- Urra H, Dufey E, Avril T, Chevet E, Hetz C, Endoplasmic reticulum stress and the hallmarks of cancer, Trends Cancer 2 (5) (2016) 252–262. - PubMed
-
- Hetz C, Papa FR, The unfolded protein response and cell fate control, Mol. Cell 69 (2) (2018) 169–181. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

