Screening of colistin-resistant bacteria in livestock animals from France
- PMID: 36414994
- PMCID: PMC9682759
- DOI: 10.1186/s13567-022-01113-1
Screening of colistin-resistant bacteria in livestock animals from France
Abstract
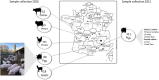
Colistin is frequently used as a growth factor or treatment against infectious bacterial diseases in animals. The Veterinary Division of the European Medicines Agency (EMA) restricted colistin use as a second-line treatment to reduce colistin resistance. In 2020, 282 faecal samples were collected from chickens, cattle, sheep, goats, and pigs in the south of France. In order to track the emergence of mobilized colistin resistant (mcr) genes in pigs, 111 samples were re-collected in 2021 and included pig faeces, food, and water from the same location. All samples were cultured in a selective Lucie Bardet Jean-Marc Rolain (LBJMR) medium and colonies were identified using MALDI-TOF mass spectrometry and then antibiotic susceptibility tests were performed. PCR and Sanger sequencing were performed to screen for the presence of mcr genes. The selective culture revealed the presence of 397 bacteria corresponding to 35 different bacterial species including Gram-negative and Gram-positive. Pigs had the highest prevalence of colistin-resistant bacteria with an abundance of intrinsically colistin-resistant bacteria and from these samples one strain harbouring both mcr-1 and mcr-3 has been isolated. The second collection allowed us to identify 304 bacteria and revealed the spread of mcr-1 and mcr-3 in pigs. In the other samples, naturally, colistin-resistant bacteria were more frequent, nevertheless the mcr-1 variant was the most abundant gene found in chicken, sheep, and goat samples and one cattle sample was positive for the mcr-3 gene. Animals are potential reservoir of colistin-resistant bacteria which varies from one animal to another. Interventions and alternative options are required to reduce the emergence of colistin resistance and to avoid zoonotic transmissions.
Keywords: France; animals; colistin-resistant bacteria; mcr genes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Kontopidou F, Plachouras D, Papadomichelakis E, Koukos G, Galani I, Poulakou G, Dimopoulos G, Antoniadou A, Armaganidis A, Giamarellou H. Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin Microbiol Infect. 2011;17:E9–E11. doi: 10.1111/j.1469-0691.2011.03649.x. - DOI - PubMed
-
- Medicines Agency E. Updated advice on the use of colistin products in animals within the European Union: development of resistance and possible impact on human and animal health. EMA. 2016;23:15–73.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

