This is a preprint.
Antiviral innate immunity is diminished in the upper respiratory tract of severe COVID-19 patients
- PMID: 36415460
- PMCID: PMC9681051
- DOI: 10.1101/2022.11.08.22281846
Antiviral innate immunity is diminished in the upper respiratory tract of severe COVID-19 patients
Abstract
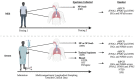
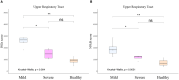
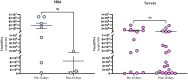
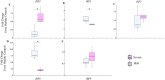
Understanding early innate immune responses to coronavirus disease 2019 (COVID-19) is crucial to developing targeted therapies to mitigate disease severity. Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 infection elicits interferon expression leading to transcription of IFN-stimulated genes (ISGs) to control viral replication and spread. SARS-CoV-2 infection also elicits NF-κB signaling which regulates inflammatory cytokine expression contributing to viral control and likely disease severity. Few studies have simultaneously characterized these two components of innate immunity to COVID-19. We designed a study to characterize the expression of interferon alpha-2 (IFNA2) and interferon beta-1 (IFNB1), both type-1 interferons (IFN-1), interferon-gamma (IFNG), a type-2 interferon (IFN-2), ISGs, and NF-κB response genes in the upper respiratory tract (URT) of patients with mild (outpatient) versus severe (hospitalized) COVID-19. Further, we characterized the weekly dynamics of these responses in the upper and lower respiratory tracts (LRTs) and blood of severe patients to evaluate for compartmental differences. We observed significantly increased ISG and NF-κB responses in the URT of mild compared with severe patients early during illness. This pattern was associated with increased IFNA2 and IFNG expression in the URT of mild patients, a trend toward increased IFNB1-expression and significantly increased STING/IRF3/cGAS expression in the URT of severe patients. Our by-week across-compartment analysis in severe patients revealed significantly higher ISG responses in the blood compared with the URT and LRT of these patients during the first week of illness, despite significantly lower expression of IFNA2, IFNB1, and IFNG in blood. NF-κB responses, however, were significantly elevated in the LRT compared with the URT and blood of severe patients during peak illness (week 2). Our data support that severe COVID-19 is associated with impaired interferon signaling in the URT during early illness and robust pro-inflammatory responses in the LRT during peak illness.
Keywords: COVID-19; Gene Expression; Inflammation; Innate Immune Response; Interferons.
Conflict of interest statement
Conflict of Interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Analysis of blood and nasal epithelial transcriptomes to identify mechanisms associated with control of SARS-CoV-2 viral load in the upper respiratory tract.J Infect. 2023 Dec;87(6):538-550. doi: 10.1016/j.jinf.2023.10.009. Epub 2023 Oct 18. J Infect. 2023. PMID: 37863321
-
Innate immune signatures in the nasopharynx after SARS-CoV-2 infection and links with the clinical outcome of COVID-19 in Omicron-dominant period.Cell Mol Life Sci. 2024 Aug 22;81(1):364. doi: 10.1007/s00018-024-05401-1. Cell Mol Life Sci. 2024. PMID: 39172244 Free PMC article.
-
Innate and Adaptive Immune Responses in the Upper Respiratory Tract and the Infectivity of SARS-CoV-2.Viruses. 2022 Apr 29;14(5):933. doi: 10.3390/v14050933. Viruses. 2022. PMID: 35632675 Free PMC article. Review.
-
Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy.Front Immunol. 2022 May 17;13:888897. doi: 10.3389/fimmu.2022.888897. eCollection 2022. Front Immunol. 2022. PMID: 35663932 Free PMC article. Review.
References
-
- Schoggins JW. Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol. 2019;6(1):567–84. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
