Thirty novel sequence variants impacting human intracranial volume
- PMID: 36415660
- PMCID: PMC9677475
- DOI: 10.1093/braincomms/fcac271
Thirty novel sequence variants impacting human intracranial volume
Abstract
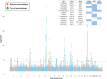
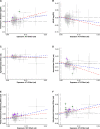
Intracranial volume, measured through magnetic resonance imaging and/or estimated from head circumference, is heritable and correlates with cognitive traits and several neurological disorders. We performed a genome-wide association study meta-analysis of intracranial volume (n = 79 174) and found 64 associating sequence variants explaining 5.0% of its variance. We used coding variation, transcript and protein levels, to uncover 12 genes likely mediating the effect of these variants, including GLI3 and CDK6 that affect cranial synostosis and microcephaly, respectively. Intracranial volume correlates genetically with volumes of cortical and sub-cortical regions, cognition, learning, neonatal and neurological traits. Parkinson's disease cases have greater and attention deficit hyperactivity disorder cases smaller intracranial volume than controls. Our Mendelian randomization studies indicate that intracranial volume associated variants either increase the risk of Parkinson's disease and decrease the risk of attention deficit hyperactivity disorder and neuroticism or correlate closely with a confounder.
Keywords: Mendelian randomization; brain structure; genetic correlation; genome-wide association study; intracranial volume.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures




References
-
- Smit DJ, Luciano M, Bartels M, et al. . Heritability of head size in Dutch and Australian twin families at ages 0–50 years. Twin Res Hum Genet. 2010;13(4):370–380. - PubMed
-
- Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. . CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505(7483):361–366. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
