Phenotyping single-cell motility in microfluidic confinement
- PMID: 36416411
- PMCID: PMC9683786
- DOI: 10.7554/eLife.76519
Phenotyping single-cell motility in microfluidic confinement
Abstract
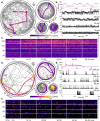
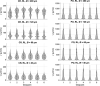
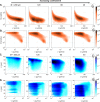
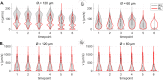
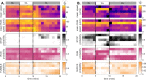
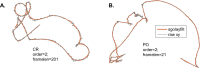
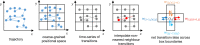
The movement trajectories of organisms serve as dynamic read-outs of their behaviour and physiology. For microorganisms this can be difficult to resolve due to their small size and fast movement. Here, we devise a novel droplet microfluidics assay to encapsulate single micron-sized algae inside closed arenas, enabling ultralong high-speed tracking of the same cell. Comparing two model species - Chlamydomonas reinhardtii (freshwater, 2 cilia), and Pyramimonas octopus (marine, 8 cilia), we detail their highly-stereotyped yet contrasting swimming behaviours and environmental interactions. By measuring the rates and probabilities with which cells transition between a trio of motility states (smooth-forward swimming, quiescence, tumbling or excitable backward swimming), we reconstruct the control network that underlies this gait switching dynamics. A simplified model of cell-roaming in circular confinement reproduces the observed long-term behaviours and spatial fluxes, including novel boundary circulation behaviour. Finally, we establish an assay in which pairs of droplets are fused on demand, one containing a trapped cell with another containing a chemical that perturbs cellular excitability, to reveal how aneural microorganisms adapt their locomotor patterns in real-time.
Keywords: algae; chlamydomonas reinhardtii; cilia; microfluidics; microswimmer; motility; physics of living systems; single cell.
© 2022, Bentley, Laeverenz-Schlogelhofer et al.
Conflict of interest statement
SB, HL, VA, JC, MM, FG, KW No competing interests declared
Figures













References
-
- Bechtold B, Fletcher P, Gorur-Shandilya S. Bastibe/violinplot-matlab: A good starting point. Zenodo. 2021 https://zenodo.org/record/4559847#.Y1loD3ZBzIU
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

