Non-cyanobacterial diazotrophs: global diversity, distribution, ecophysiology, and activity in marine waters
- PMID: 36416813
- PMCID: PMC10719068
- DOI: 10.1093/femsre/fuac046
Non-cyanobacterial diazotrophs: global diversity, distribution, ecophysiology, and activity in marine waters
Abstract
Biological dinitrogen (N2) fixation supplies nitrogen to the oceans, supporting primary productivity, and is carried out by some bacteria and archaea referred to as diazotrophs. Cyanobacteria are conventionally considered to be the major contributors to marine N2 fixation, but non-cyanobacterial diazotrophs (NCDs) have been shown to be distributed throughout ocean ecosystems. However, the biogeochemical significance of marine NCDs has not been demonstrated. This review synthesizes multiple datasets, drawing from cultivation-independent molecular techniques and data from extensive oceanic expeditions, to provide a comprehensive view into the diversity, biogeography, ecophysiology, and activity of marine NCDs. A NCD nifH gene catalog was compiled containing sequences from both PCR-based and PCR-free methods, identifying taxa for future studies. NCD abundances from a novel database of NCD nifH-based abundances were colocalized with environmental data, unveiling distinct distributions and environmental drivers of individual taxa. Mechanisms that NCDs may use to fuel and regulate N2 fixation in response to oxygen and fixed nitrogen availability are discussed, based on a metabolic analysis of recently available Tara Oceans expedition data. The integration of multiple datasets provides a new perspective that enhances understanding of the biology, ecology, and biogeography of marine NCDs and provides tools and directions for future research.
Keywords: diazotrophs; marine nitrogen cycle; nitrogen fixation; non-cyanobacterial diazotrophs.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures
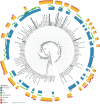

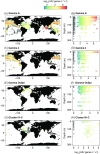
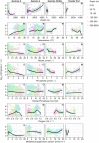



References
-
- Ashkezari MD, Hagen NR, Denholtz Met al. . Simons Collaborative Marine Atlas Project (Simons CMAP): an open-source portal to share, visualize, and analyze ocean data. Limnol Oceanogr Methods. 2021;19:488–96.
-
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. - PubMed
-
- Benavides M, Bonnet S, Berman-Frank Iet al. . Deep into oceanic N2 fixation. Front Mar Sci. 2018a;5:1–4. - PubMed

