PHOSPHATE RESPONSE 1 family members act distinctly to regulate transcriptional responses to phosphate starvation
- PMID: 36417239
- PMCID: PMC9922430
- DOI: 10.1093/plphys/kiac521
PHOSPHATE RESPONSE 1 family members act distinctly to regulate transcriptional responses to phosphate starvation
Abstract
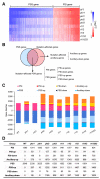
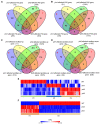
To sustain growth when facing phosphate (Pi) starvation, plants trigger an array of adaptive responses that are largely controlled at transcriptional levels. In Arabidopsis (Arabidopsis thaliana), the four transcription factors of the PHOSPHATE RESPONSE 1 (PHR1) family, PHR1 and its homologs PHR1-like 1 (PHL1), PHL2, and PHL3 form the central regulatory system that controls the expression of Pi starvation-responsive (PSR) genes. However, how each of these four proteins function in regulating the transcription of PSR genes remains largely unknown. In this work, we performed comparative phenotypic and transcriptomic analyses using Arabidopsis mutants with various combinations of mutations in these four genes. The results showed that PHR1/PHL1 and PHL2/PHL3 do not physically interact with each other and function as two distinct modules in regulating plant development and transcriptional responses to Pi starvation. In the PHR1/PHL1 module, PHR1 plays a dominant role, whereas, in the PHL2/PHL3 module, PHL2 and PHL3 contribute similarly to the regulation of PSR gene transcription. By analyzing their common and specific targets, we showed that these PHR proteins could function as both positive and negative regulators of PSR gene expression depending on their targets. Some interactions between PHR1 and PHL2/PHL3 in regulating PSR gene expression were also observed. In addition, we identified a large set of defense-related genes whose expression is not affected in wild-type plants but is altered in the mutant plants under Pi starvation. These results increase our understanding of the molecular mechanism underlying plant transcriptional responses to Pi starvation.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare that there was no conflict of interest.
Figures






References
-
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8): 1194–1202 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

