The GGDEF-EAL protein CdgB from Azospirillum baldaniorum Sp245, is a dual function enzyme with potential polar localization
- PMID: 36417483
- PMCID: PMC9683572
- DOI: 10.1371/journal.pone.0278036
The GGDEF-EAL protein CdgB from Azospirillum baldaniorum Sp245, is a dual function enzyme with potential polar localization
Abstract
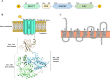
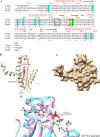
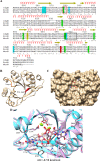
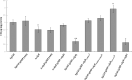
Azospirillum baldaniorum Sp245, a plant growth-promoting rhizobacterium, can form biofilms through a process controlled by the second messenger cyclic diguanylate monophosphate (c-di-GMP). A. baldaniorum has a variety of proteins potentially involved in controlling the turnover of c-di-GMP many of which are coupled to sensory domains that could be involved in establishing a mutualistic relationship with the host. Here, we present in silico analysis and experimental characterization of the function of CdgB (AZOBR_p410089), a predicted MHYT-PAS-GGDEF-EAL multidomain protein from A. baldaniorum Sp245. When overproduced, CdgB behaves predominantly as a c-di-GMP phosphodiesterase (PDE) in A. baldaniorum Sp245. It inhibits biofilm formation and extracellular polymeric substances production and promotes swimming motility. However, a CdgB variant with a degenerate PDE domain behaves as diguanylate cyclase (DGC). This strongly suggest that CdgB is capable of dual activity. Variants with alterations in the DGC domain and the MHYT domain negatively affects extracellular polymeric substances production and induction of swimming motility. Surprisingly, we observed that overproduction of CdgB results in increased c-di-GMP accumulation in the heterologous host Escherichia coli, suggesting under certain conditions, the WT CdgB variant can behave predominantly as a DGC. Furthermore, we also demonstrated that CdgB is anchored to the cell membrane and localizes potentially to the cell poles. This localization is dependent on the presence of the MHYT domain. In summary, our results suggest that CdgB can provide versatility to signaling modules that control motile and sessile lifestyles in response to key environmental signals in A. baldaniorum.
Copyright: © 2022 Viruega-Góngora et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
In silico comparative analysis of GGDEF and EAL domain signaling proteins from the Azospirillum genomes.BMC Microbiol. 2018 Mar 9;18(1):20. doi: 10.1186/s12866-018-1157-0. BMC Microbiol. 2018. PMID: 29523074 Free PMC article.
-
The dual GGDEF/EAL domain enzyme PA0285 is a Pseudomonas species housekeeping phosphodiesterase regulating early attachment and biofilm architecture.J Biol Chem. 2024 Feb;300(2):105659. doi: 10.1016/j.jbc.2024.105659. Epub 2024 Jan 16. J Biol Chem. 2024. PMID: 38237678 Free PMC article.
-
GepA, a GGDEF-EAL protein, regulates biofilm formation and swimming motility in Vibrio parahaemolyticus.Arch Microbiol. 2025 Mar 22;207(5):99. doi: 10.1007/s00203-025-04282-7. Arch Microbiol. 2025. PMID: 40119885
-
Progress in Understanding the Molecular Basis Underlying Functional Diversification of Cyclic Dinucleotide Turnover Proteins.J Bacteriol. 2017 Feb 14;199(5):e00790-16. doi: 10.1128/JB.00790-16. Print 2017 Mar 1. J Bacteriol. 2017. PMID: 28031279 Free PMC article. Review.
-
Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins.Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150498. doi: 10.1098/rstb.2015.0498. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672149 Free PMC article. Review.
Cited by
-
Stress adaptation under in vitro evolution influences survival and metabolic phenotypes of clinical and environmental strains of Vibrio cholerae El-Tor.Microbiol Spectr. 2025 Mar 4;13(3):e0121124. doi: 10.1128/spectrum.01211-24. Epub 2025 Feb 11. Microbiol Spectr. 2025. PMID: 39932327 Free PMC article.
-
Cyclic Diguanylate in the Wild: Roles During Plant and Animal Colonization.Annu Rev Microbiol. 2024 Nov;78(1):533-551. doi: 10.1146/annurev-micro-041522-101729. Epub 2024 Nov 7. Annu Rev Microbiol. 2024. PMID: 39270684 Free PMC article. Review.
-
Characterization of a MHYT domain-coupled transcriptional regulator that responds to carbon monoxide.Nucleic Acids Res. 2024 Aug 27;52(15):8849-8860. doi: 10.1093/nar/gkae575. Nucleic Acids Res. 2024. PMID: 38966994 Free PMC article.
-
Functional analysis of cyclic diguanylate-modulating proteins in Vibrio fischeri.mSystems. 2024 Nov 19;9(11):e0095624. doi: 10.1128/msystems.00956-24. Epub 2024 Oct 22. mSystems. 2024. PMID: 39436151 Free PMC article.
-
Functional analysis of cyclic diguanylate-modulating proteins in Vibrio fischeri.bioRxiv [Preprint]. 2024 Sep 24:2023.07.24.550417. doi: 10.1101/2023.07.24.550417. bioRxiv. 2024. Update in: mSystems. 2024 Nov 19;9(11):e0095624. doi: 10.1128/msystems.00956-24. PMID: 37546929 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

