Reprogramming Müller glia to regenerate ganglion-like cells in adult mouse retina with developmental transcription factors
- PMID: 36417510
- PMCID: PMC9683702
- DOI: 10.1126/sciadv.abq7219
Reprogramming Müller glia to regenerate ganglion-like cells in adult mouse retina with developmental transcription factors
Erratum in
-
Erratum for the Research Article: "Reprogramming Müller glia to regenerate ganglion-like cells in adult mouse retina with developmental transcription factors".Sci Adv. 2023 Jun 9;9(23):eadi5564. doi: 10.1126/sciadv.adi5564. Epub 2023 Jun 9. Sci Adv. 2023. PMID: 37294771 Free PMC article. No abstract available.
Abstract
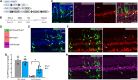
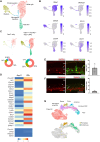
Many neurodegenerative diseases cause degeneration of specific types of neurons. For example, glaucoma leads to death of retinal ganglion cells, leaving other neurons intact. Neurons are not regenerated in the adult mammalian central nervous system. However, in nonmammalian vertebrates, glial cells spontaneously reprogram into neural progenitors and replace neurons after injury. We have recently developed strategies to stimulate regeneration of functional neurons in the adult mouse retina by overexpressing the proneural factor Ascl1 in Müller glia. Here, we test additional transcription factors (TFs) for their ability to direct regeneration to particular types of retinal neurons. We engineered mice to express different combinations of TFs in Müller glia, including Ascl1, Pou4f2, Islet1, and Atoh1. Using immunohistochemistry, single-cell RNA sequencing, single-cell assay for transposase-accessible chromatin sequencing, and electrophysiology, we find that retinal ganglion-like cells can be regenerated in the damaged adult mouse retina in vivo with targeted overexpression of developmental retinal ganglion cell TFs.
Figures





References
-
- A. Bringmann, I. Iandiev, T. Pannicke, A. Wurm, M. Hollborn, P. Wiedemann, N. N. Osborne, A. Reichenbach,Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Prog. Retin. Eye Res. 28,423–451 (2009). - PubMed
-
- N. L. Jorstad, M. S. Wilken, L. Todd, C. Finkbeiner, P. Nakamura, N. Radulovich, M. J. Hooper, A. Chitsazan, B. A. Wilkerson, F. Rieke, T. A. Reh,STAT signaling modifies Ascl1 chromatin binding and limits neural regeneration from Muller glia in adult mouse retina. Cell Rep. 30,2195–2208.e5 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

