Integration of 3D genome topology and local chromatin features uncovers enhancers underlying craniofacial-specific cartilage defects
- PMID: 36417512
- PMCID: PMC9683718
- DOI: 10.1126/sciadv.abo3648
Integration of 3D genome topology and local chromatin features uncovers enhancers underlying craniofacial-specific cartilage defects
Abstract
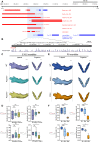
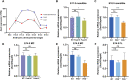
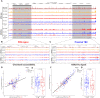
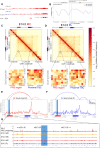
Aberrations in tissue-specific enhancers underlie many developmental defects. Disrupting a noncoding region distal from the human SOX9 gene causes the Pierre Robin sequence (PRS) characterized by the undersized lower jaw. Such a craniofacial-specific defect has been previously linked to enhancers transiently active in cranial neural crest cells (CNCCs). We demonstrate that the PRS region also strongly regulates Sox9 in CNCC-derived Meckel's cartilage (MC), but not in limb cartilages, even after decommissioning of CNCC enhancers. Such an MC-specific regulatory effect correlates with the MC-specific chromatin contacts between the PRS region and Sox9, highlighting the importance of lineage-dependent chromatin topology in instructing enhancer usage. By integrating the enhancer signatures and chromatin topology, we uncovered >10,000 enhancers that function differentially between MC and limb cartilages and demonstrated their association with human diseases. Our findings provide critical insights for understanding the choreography of gene regulation during development and interpreting the genetic basis of craniofacial pathologies.
Figures
Similar articles
-
Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence.Hum Mutat. 2014 Aug;35(8):1011-20. doi: 10.1002/humu.22606. Hum Mutat. 2014. PMID: 24934569 Free PMC article.
-
Loss of Extreme Long-Range Enhancers in Human Neural Crest Drives a Craniofacial Disorder.Cell Stem Cell. 2020 Nov 5;27(5):765-783.e14. doi: 10.1016/j.stem.2020.09.001. Epub 2020 Sep 28. Cell Stem Cell. 2020. PMID: 32991838 Free PMC article.
-
Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence.Nat Genet. 2009 Mar;41(3):359-64. doi: 10.1038/ng.329. Epub 2009 Feb 22. Nat Genet. 2009. PMID: 19234473
-
Human genetic variation within neural crest enhancers: molecular and phenotypic implications.Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120360. doi: 10.1098/rstb.2012.0360. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650634 Free PMC article. Review.
-
Developmental and genetic perspectives on Pierre Robin sequence.Am J Med Genet C Semin Med Genet. 2013 Nov;163C(4):295-305. doi: 10.1002/ajmg.c.31374. Epub 2013 Oct 11. Am J Med Genet C Semin Med Genet. 2013. PMID: 24127256 Review.
Cited by
-
ARID1A safeguards the canalization of the cell fate decision during osteoclastogenesis.Nat Commun. 2024 Jul 17;15(1):5994. doi: 10.1038/s41467-024-50225-z. Nat Commun. 2024. PMID: 39013863 Free PMC article.
-
A mathematical model clarifies the ABC Score formula used in enhancer-gene prediction.bioRxiv [Preprint]. 2024 Dec 3:2024.11.29.626072. doi: 10.1101/2024.11.29.626072. bioRxiv. 2024. PMID: 39677755 Free PMC article. Preprint.
-
Cis-Regulatory Elements in Mammals.Int J Mol Sci. 2023 Dec 26;25(1):343. doi: 10.3390/ijms25010343. Int J Mol Sci. 2023. PMID: 38203513 Free PMC article. Review.
-
SATB1 prevents immune cell infiltration by regulating chromatin organization and gene expression of a chemokine gene cluster in T cells.Commun Biol. 2024 Oct 11;7(1):1304. doi: 10.1038/s42003-024-07021-8. Commun Biol. 2024. PMID: 39394451 Free PMC article.
-
Shaping faces: genetic and epigenetic control of craniofacial morphogenesis.Nat Rev Genet. 2023 Sep;24(9):610-626. doi: 10.1038/s41576-023-00594-w. Epub 2023 Apr 24. Nat Rev Genet. 2023. PMID: 37095271 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

