Multi-omics analysis identifies osteosarcoma subtypes with distinct prognosis indicating stratified treatment
- PMID: 36418292
- PMCID: PMC9684515
- DOI: 10.1038/s41467-022-34689-5
Multi-omics analysis identifies osteosarcoma subtypes with distinct prognosis indicating stratified treatment
Abstract
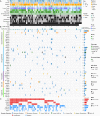
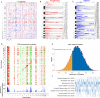
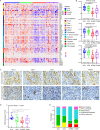
Osteosarcoma (OS) is a primary malignant bone tumor that most commonly affects children, adolescents, and young adults. Here, we comprehensively analyze genomic, epigenomic and transcriptomic data from 121 OS patients. Somatic mutations are diverse within the cohort, and only TP53 is significantly mutated. Through unsupervised integrative clustering of the multi-omics data, we classify OS into four subtypes with distinct molecular features and clinical prognosis: (1) Immune activated (S-IA), (2) Immune suppressed (S-IS), (3) Homologous recombination deficiency dominant (S-HRD), and (4) MYC driven (S-MD). MYC amplification with HR proficiency tumors is identified with a high oxidative phosphorylation signature resulting in resistance to neoadjuvant chemotherapy. Potential therapeutic targets are identified for each subtype, including platinum-based chemotherapy, immune checkpoint inhibitors, anti-VEGFR, anti-MYC and PARPi-based synthetic lethal strategies. Our comprehensive integrated characterization provides a valuable resource that deepens our understanding of the disease, and may guide future clinical strategies for the precision treatment of OS.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- George S. Developments in systemic therapy for soft tissue and bone sarcomas. J. Natl Compr. Canc. Netw. 2019;17:625–628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

