Sphingosine 1-phosphate mediates adiponectin receptor signaling essential for lipid homeostasis and embryogenesis
- PMID: 36418331
- PMCID: PMC9684441
- DOI: 10.1038/s41467-022-34931-0
Sphingosine 1-phosphate mediates adiponectin receptor signaling essential for lipid homeostasis and embryogenesis
Abstract
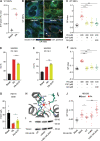
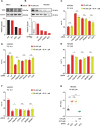
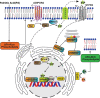
Cells and organisms require proper membrane composition to function and develop. Phospholipids are the major component of membranes and are primarily acquired through the diet. Given great variability in diet composition, cells must be able to deploy mechanisms that correct deviations from optimal membrane composition and properties. Here, using lipidomics and unbiased proteomics, we found that the embryonic lethality in mice lacking the fluidity regulators Adiponectin Receptors 1 and 2 (AdipoR1/2) is associated with aberrant high saturation of the membrane phospholipids. Using mouse embryonic fibroblasts (MEFs) derived from AdipoR1/2-KO embryos, human cell lines and the model organism C. elegans we found that, mechanistically, AdipoR1/2-derived sphingosine 1-phosphate (S1P) signals in parallel through S1PR3-SREBP1 and PPARγ to sustain the expression of the fatty acid desaturase SCD and maintain membrane properties. Thus, our work identifies an evolutionary conserved pathway by which cells and organisms achieve membrane homeostasis and adapt to a variable environment.
© 2022. The Author(s).
Conflict of interest statement
K.P. and M.B. are presently employed by AstraZeneca and may be AstraZeneca shareholders. The remaining others declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

