High-precision estimation of emitter positions using Bayesian grouping of localizations
- PMID: 36418347
- PMCID: PMC9684143
- DOI: 10.1038/s41467-022-34894-2
High-precision estimation of emitter positions using Bayesian grouping of localizations
Abstract
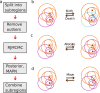
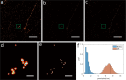
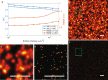
Single-molecule localization microscopy super-resolution methods rely on stochastic blinking/binding events, which often occur multiple times from each emitter over the course of data acquisition. Typically, the blinking/binding events from each emitter are treated as independent events, without an attempt to assign them to a particular emitter. Here, we describe a Bayesian method of inferring the positions of the tagged molecules by exploring the possible grouping and combination of localizations from multiple blinking/binding events. The results are position estimates of the tagged molecules that have improved localization precision and facilitate nanoscale structural insights. The Bayesian framework uses the localization precisions to learn the statistical distribution of the number of blinking/binding events per emitter and infer the number and position of emitters. We demonstrate the method on a range of synthetic data with various emitter densities, DNA origami constructs and biological structures using DNA-PAINT and dSTORM data. We show that under some experimental conditions it is possible to achieve sub-nanometer precision.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

