Inflammatory bone marrow signaling in pediatric acute myeloid leukemia distinguishes patients with poor outcomes
- PMID: 36418348
- PMCID: PMC9684530
- DOI: 10.1038/s41467-022-34965-4
Inflammatory bone marrow signaling in pediatric acute myeloid leukemia distinguishes patients with poor outcomes
Abstract
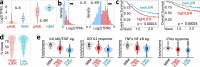
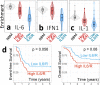
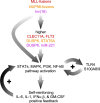
High levels of the inflammatory cytokine IL-6 in the bone marrow are associated with poor outcomes in pediatric acute myeloid leukemia (pAML), but its etiology remains unknown. Using RNA-seq data from pre-treatment bone marrows of 1489 children with pAML, we show that > 20% of patients have concurrent IL-6, IL-1, IFNα/β, and TNFα signaling activity and poorer outcomes. Targeted sequencing of pre-treatment bone marrow samples from affected patients (n = 181) revealed 5 highly recurrent patterns of somatic mutation. Using differential expression analyses of the most common genomic subtypes (~60% of total), we identify high expression of multiple potential drivers of inflammation-related treatment resistance. Regardless of genomic subtype, we show that JAK1/2 inhibition reduces receptor-mediated inflammatory signaling by leukemic cells in-vitro. The large number of high-risk pAML genomic subtypes presents an obstacle to the development of mutation-specific therapies. Our findings suggest that therapies targeting inflammatory signaling may be effective across multiple genomic subtypes of pAML.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

