Transcriptome analysis of aphids exposed to glandular trichomes in tomato reveals stress and starvation related responses
- PMID: 36418431
- PMCID: PMC9684535
- DOI: 10.1038/s41598-022-24490-1
Transcriptome analysis of aphids exposed to glandular trichomes in tomato reveals stress and starvation related responses
Abstract
Understanding the responses of insect herbivores to plant chemical defences is pivotal for the management of crops and pests. However, the mechanisms of interaction are not entirely understood. In this study, we compared the whole transcriptome gene expression of the aphid Macrosiphum euphorbiae grown on two different varieties of tomato that differ in their inducible chemical defences. We used two isogenic lines of tomato with a shared genetic background that only differ in the presence of type IV glandular trichomes and their associated acylsucrose excretions. This works also reports a de novo transcriptome of the aphid M. euphorbiae. Subsequently, we identified a unique and distinct gene expression profile for the first time corresponding to aphid´s exposure to type IV glandular trichomes and acylsugars. The analysis of the aphid transcriptome shows that tomato glandular trichomes and their associated secretions are highly efficient in triggering stress-related responses in the aphid, and demonstrating that their role in plant defence goes beyond the physical impediment of herbivore activity. Some of the differentially expressed genes were associated with carbohydrate, lipid and xenobiotic metabolisms, immune system, oxidative stress response and hormone biosynthesis pathways. Also, the observed responses are compatible with a starvation syndrome. The transcriptome analysis puts forward a wide range of genes involved in the synthesis and regulation of detoxification enzymes that reveal important underlying mechanisms in the interaction of the aphid with its host plant and provides a valuable genomic resource for future study of biological processes at the molecular level using this aphid.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
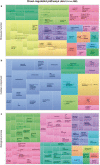
Figures





References
-
- Ehrlich, P. R. & Raven, P. H. Butterflies and plants: a study in coevolution. Evolution (N Y)18, (1964).
-
- Hembry, D. H., Yoder, J. B. & Goodman, K. R. Coevolution and the diversification of life. Am Nat184, (2014). - PubMed
-
- Nishida, R. Sequestration of defensive substances from plants by Lepidoptera. Annu. Rev. Entomol. vol. 47 Preprint at 10.1146/annurev.ento.47.091201.145121 (2002). - PubMed
-
- Rathcke, B. J. & Poole, R. W. Coevolutionary race continues: butterfly larval adaptation to plant trichomes. Science (1979)187, (1975). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

