Protein conformation stabilized by newly formed turns for thermal resilience
- PMID: 36419349
- PMCID: PMC9822789
- DOI: 10.1016/j.bpj.2022.11.2936
Protein conformation stabilized by newly formed turns for thermal resilience
Abstract
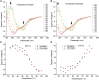
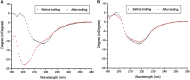
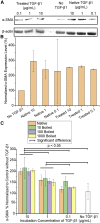
Thermally stable or resilient proteins are usually stabilized at intermediate states during thermal stress to prevent irreversible denaturation. However, the mechanism by which their conformations are stabilized to resist high temperature remains elusive. Herein, we investigate the conformational and thermal stability of transforming growth factor-β1 (TGF-β1), a key signaling molecule in numerous biological pathways. We report that the TGF-β1 molecule is thermally resilient as it gradually denatures during thermal treatment when the temperature increases to 90°C-100°C but recovers native folding when the temperature decreases. Using this protein as a model, further studies show the maintenance of its bioactive functional properties after thermal stress, as demonstrated by differentiation induction of NIH/3T3 fibroblasts and human mesenchymal stem cells into myofibroblasts and chondrocytes, respectively. Molecular dynamic simulations revealed that although the protein's secondary structure is unstable under thermal stress, its conformation is partially stabilized by newly formed turns. Given the importance and/or prevalence of TGF-β1 in biological processes, potential therapeutics, and the human diet, our findings encourage consideration of its thermostability for biomedical applications and nutrition.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Vogt G., Argos P. Protein thermal stability: hydrogen bonds or internal packing? Folding Des. 1997;2:S40–S46. - PubMed
-
- Simpson S., Kaislasuo J., et al. Pal L. Thermal stability of cytokines: a review. Cytokine. 2020;125:154829. - PubMed
-
- Leuenberger P., Ganscha S., et al. Picotti P. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355:eaai7825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

