Population-wide gene disruption in the murine lung epithelium via AAV-mediated delivery of CRISPR-Cas9 components
- PMID: 36419469
- PMCID: PMC9668740
- DOI: 10.1016/j.omtm.2022.10.016
Population-wide gene disruption in the murine lung epithelium via AAV-mediated delivery of CRISPR-Cas9 components
Abstract
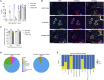
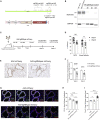
With the aim of expediting drug target discovery and validation for respiratory diseases, we developed an optimized method for in situ somatic gene disruption in murine lung epithelial cells via AAV6-mediated CRISPR-Cas9 delivery. Efficient gene editing was observed in lung type II alveolar epithelial cells and distal airway cells following assessment of single- or dual-guide AAV vector formats, Cas9 variants, and a sequential dosing strategy with combinatorial guide RNA expression cassettes. In particular, we were able to demonstrate population-wide gene disruption within distinct epithelial cell types for separate targets in Cas9 transgenic animals, with minimal to no associated inflammation. We also observed and characterized AAV vector integration events that occurred within directed double-stranded DNA break sites in lung cells, highlighting a complicating factor with AAV-mediated delivery of DNA nucleases. Taken together, we demonstrate a uniquely effective approach for somatic engineering of the murine lung, which will greatly facilitate the modeling of disease and therapeutic intervention.
Keywords: AAV; CRISPR; Cas9; adeno-associated virus; disease modeling; gene editing; genome integration; inflammatory response; lung epithelium; viral delivery.
© 2022 The Author(s).
Conflict of interest statement
During the course of these studies, all authors were full-time employees of Genentech, Inc., and potential shareholders of Roche. No other disclosures were reported.
Figures




References
-
- Pacak C.A., Mah C.S., Thattaliyath B.D., Conlon T.J., Lewis M.A., Cloutier D.E., Zolotukhin I., Tarantal A.F., Byrne B.J. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 2006;99:e3–e9. doi: 10.1161/01.res.0000237661.18885.f6. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

