Phosphorylation of T897 in the dimerization domain of Gemin5 modulates protein interactions and translation regulation
- PMID: 36420152
- PMCID: PMC9676205
- DOI: 10.1016/j.csbj.2022.11.018
Phosphorylation of T897 in the dimerization domain of Gemin5 modulates protein interactions and translation regulation
Abstract
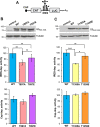
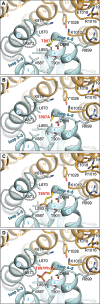
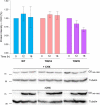
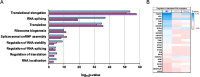
Gemin5 is a multifunctional RNA binding protein (RBP) organized in domains with a distinctive structural organization. The protein is a hub for several protein networks performing diverse RNA-dependent functions including regulation of translation, and recognition of small nuclear RNAs (snRNAs). Here we sought to identify the presence of phosphoresidues on the C-terminal half of Gemin5, a region of the protein that harbors a tetratricopeptide repeat (TPR)-like dimerization domain and a non-canonical RNA binding site (RBS1). We identified two phosphoresidues in the purified protein: P-T897 in the dimerization domain and P-T1355 in RBS1. Replacing T897 and T1355 with alanine led to decreased translation, and mass spectrometry analysis revealed that mutation T897A strongly abrogates the association with cellular proteins related to the regulation of translation. In contrast, the phosphomimetic substitutions to glutamate partially rescued the translation regulatory activity. The structural analysis of the TPR dimerization domain indicates that local rearrangements caused by phosphorylation of T897 affect the conformation of the flexible loop 2-3, and propagate across the dimerization interface, impacting the position of the C-terminal helices and the loop 12-13 shown to be mutated in patients with neurological disorders. Computational analysis of the potential relationship between post-translation modifications and currently known pathogenic variants indicates a lack of overlapping of the affected residues within the functional domains of the protein and provides molecular insights for the implication of the phosphorylated residues in translation regulation.
Keywords: BiNGO, Biological Networks Gene Ontology application; CHX, cycloheximide; Gemin5 interactome; Human variants; IRES, internal ribosome entry site; LC-MS/MS, liquid chromatography-mass spectrometry; MD, molecular dynamics; NEDCAM, neurological disorders with cerebellar atrophy and motor dysfunction; Neurological disease; Phosphoresidues; Protein structure modeling; Protein synthesis; RBP, RNA-binding protein; RBS1, RNA-binding site1; RNA-binding proteins; SGs, stress granules; SMN complex; SMN, survival of motor neurons; TAP, tandem affinity purification; TPR-like, tetratricopeptide repeat-like domain; WD-40, tryptophan-aspartic repeat motif; eIF4E, eukaryotic initiation factor 4E; snRNAs, small nuclear RNAs.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Understanding GEMIN5 Interactions: From Structural and Functional Insights to Selective Translation.Wiley Interdiscip Rev RNA. 2025 Mar-Apr;16(2):e70008. doi: 10.1002/wrna.70008. Wiley Interdiscip Rev RNA. 2025. PMID: 40176294 Free PMC article. Review.
-
Oligomerization regulates the interaction of Gemin5 with members of the SMN complex and the translation machinery.Cell Death Discov. 2024 Jun 28;10(1):306. doi: 10.1038/s41420-024-02057-5. Cell Death Discov. 2024. PMID: 38942768 Free PMC article.
-
Functional and structural deficiencies of Gemin5 variants associated with neurological disorders.Life Sci Alliance. 2022 Apr 7;5(7):e202201403. doi: 10.26508/lsa.202201403. Print 2022 Jul. Life Sci Alliance. 2022. PMID: 35393353 Free PMC article.
-
RNA-protein coevolution study of Gemin5 uncovers the role of the PXSS motif of RBS1 domain for RNA binding.RNA Biol. 2020 Sep;17(9):1331-1341. doi: 10.1080/15476286.2020.1762054. Epub 2020 May 31. RNA Biol. 2020. PMID: 32476560 Free PMC article.
-
Gemin5: A Multitasking RNA-Binding Protein Involved in Translation Control.Biomolecules. 2015 Apr 17;5(2):528-44. doi: 10.3390/biom5020528. Biomolecules. 2015. PMID: 25898402 Free PMC article. Review.
Cited by
-
Function and dysfunction of GEMIN5: understanding a novel neurodevelopmental disorder.Neural Regen Res. 2024 Nov 1;19(11):2377-2386. doi: 10.4103/NRR.NRR-D-23-01614. Epub 2024 Jan 31. Neural Regen Res. 2024. PMID: 38526274 Free PMC article.
-
Understanding GEMIN5 Interactions: From Structural and Functional Insights to Selective Translation.Wiley Interdiscip Rev RNA. 2025 Mar-Apr;16(2):e70008. doi: 10.1002/wrna.70008. Wiley Interdiscip Rev RNA. 2025. PMID: 40176294 Free PMC article. Review.
-
Impact of Gemin5 in protein synthesis: phosphoresidues of the dimerization domain regulate ribosome binding.RNA Biol. 2025 Dec;22(1):1-15. doi: 10.1080/15476286.2025.2540654. Epub 2025 Aug 3. RNA Biol. 2025. PMID: 40734649 Free PMC article.
-
Expanding the clinical phenotype and genetic spectrum of GEMIN5 disorders: Early-infantile developmental and epileptic encephalopathies.Brain Behav. 2024 May;14(5):e3535. doi: 10.1002/brb3.3535. Brain Behav. 2024. PMID: 38773790 Free PMC article.
-
Oligomerization regulates the interaction of Gemin5 with members of the SMN complex and the translation machinery.Cell Death Discov. 2024 Jun 28;10(1):306. doi: 10.1038/s41420-024-02057-5. Cell Death Discov. 2024. PMID: 38942768 Free PMC article.
References
-
- Lau C.K., Bachorik J.L., Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol. 2009;16:486–491. - PubMed
-
- Fischer U., Englbrecht C., Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA. 2011;2(5):718–731. - PubMed
-
- Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem. 2007;282:5825–5833. - PubMed
-
- Battle D.J., Lau C.K., Wan L., Deng H., Lotti F., Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23:273–279. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
