Recognition of the ligand-induced spatiotemporal residue pair pattern of β2-adrenergic receptors using 3-D residual networks trained by the time series of protein distance maps
- PMID: 36420156
- PMCID: PMC9677134
- DOI: 10.1016/j.csbj.2022.10.036
Recognition of the ligand-induced spatiotemporal residue pair pattern of β2-adrenergic receptors using 3-D residual networks trained by the time series of protein distance maps
Abstract
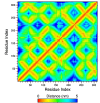
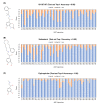
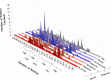
G protein-coupled receptors (GPCRs) are promising drug targets because they play a large role in physiological processes by modulating diverse signaling pathways in the human body. The GPCR-mediated signaling pathways are regulated by four types of ligands-agonists, neutral antagonists, partial agonists, and inverse agonists. Once each type of ligand is bound to the binding site, it activates, deactivates, or does not perturb signaling by shifting the conformational ensemble of GPCRs. Predicting the ligand's effect on the conformation at the binding moment could be a powerful screening tool for rational GPCR drug design. Here, we detected conformational differences by capturing the spatiotemporal residue pair pattern of the ligand-bound β2-adrenergic receptor (β2AR) using a 3-dimensional residual network, 3D-ResNets. The network was trained with the time series of protein distance maps extracted from hundreds of molecular dynamics (MD) simulation trajectories of ten β2AR-ligand complexes. The MD system was constructed with a lipid bilayer embedded in an inactive β2AR X-ray crystal structure and solvated with explicit water molecules. To train the network, three hyperparameters were tested, and it was found that the number of MD trajectories in the training set significantly affected the model's accuracy. The classification of agonists and neutral antagonists was successful, but inverse agonists were not. Between the agonists and antagonists, different residue pair patterns were spotted on the extracellular loop segment. This result demonstrates the potential application of a 3-D neural network in GPCR drug screening, as well as an analysis tool for protein functional dynamics.
Keywords: 3-D Convolution Neural Network; 3D-ResNets, 3-dimensional residual networks; Artificial Intelligence; ECL, extracellular loop; GPCR; GPCRs, G protein-coupled receptors; ICL, intracellular loop; MD, molecular dynamics; Machine Learning; Molecular Dynamics Simulation; PDM, protein distance map; Pattern Recognition; TM, TMtransmembrane helix; β2-Adrenergic Receptor; β2AR, β2-adrenergic receptor.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures











Similar articles
-
GDP Release from the Open Conformation of Gα Requires Allosteric Signaling from the Agonist-Bound Human β2 Adrenergic Receptor.J Chem Inf Model. 2020 Aug 24;60(8):4064-4075. doi: 10.1021/acs.jcim.0c00432. Epub 2020 Aug 11. J Chem Inf Model. 2020. PMID: 32786510
-
Ligand-binding affinity of alternative conformers of human β2 -adrenergic receptor in the presence of intracellular loop 3 (ICL3) and their potential use in virtual screening studies.Chem Biol Drug Des. 2019 May;93(5):883-899. doi: 10.1111/cbdd.13478. Epub 2019 Feb 12. Chem Biol Drug Des. 2019. PMID: 30637937
-
Structure-Based Prediction of G-Protein-Coupled Receptor Ligand Function: A β-Adrenoceptor Case Study.J Chem Inf Model. 2015 May 26;55(5):1045-61. doi: 10.1021/acs.jcim.5b00066. Epub 2015 May 1. J Chem Inf Model. 2015. PMID: 25848966
-
Structure and dynamics of G-protein coupled receptors.Adv Exp Med Biol. 2014;796:37-54. doi: 10.1007/978-94-007-7423-0_3. Adv Exp Med Biol. 2014. PMID: 24158800 Review.
-
Efficiency of Homology Modeling Assisted Molecular Docking in G-protein Coupled Receptors.Curr Top Med Chem. 2021;21(4):269-294. doi: 10.2174/1568026620666200908165250. Curr Top Med Chem. 2021. PMID: 32901584 Review.
Cited by
-
Deep learning for discriminating non-trivial conformational changes in molecular dynamics simulations of SARS-CoV-2 spike-ACE2.Sci Rep. 2024 Sep 30;14(1):22639. doi: 10.1038/s41598-024-72842-w. Sci Rep. 2024. PMID: 39349594 Free PMC article.
References
-
- Lohse M.J., Nuber S., Hoffmann C. Fluorescence / Bioluminescence Resonance Energy Transfer Techniques to Study G-Protein-Coupled. Pharmacol Rev. 2012;64:299–336. - PubMed
LinkOut - more resources
Full Text Sources
