Virulence-related regulatory network of Pseudomonas syringae
- PMID: 36420163
- PMCID: PMC9678800
- DOI: 10.1016/j.csbj.2022.11.011
Virulence-related regulatory network of Pseudomonas syringae
Abstract
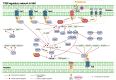
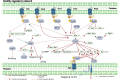
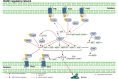
Transcription factors (TFs) play important roles in regulating multiple biological processes by binding to promoter regions and regulating the global gene transcription levels. Pseudomonas syringae is a Gram-negative phytopathogenic bacterium harbouring 301 putative TFs in its genome, approximately 50 of which are responsible for virulence-related gene and pathway regulation. Over the past decades, RNA sequencing, chromatin immunoprecipitation sequencing, high-throughput systematic evolution of ligands by exponential enrichment, and other technologies have been applied to identify the functions of master regulators and their interactions in virulence-related pathways. This review summarises the recent advances in the regulatory networks of TFs involved in the type III secretion system (T3SS) and non-T3SS virulence-associated pathways, including motility, biofilm formation, quorum sensing, nucleotide-based secondary messengers, phytotoxins, siderophore production, and oxidative stress. Moreover, this review discusses the future perspectives in terms of TF-mediated pathogenesis mechanisms and provides novel insights that will help combat P. syringae infections based on the regulatory networks of TFs.
Keywords: Pseudomonas syringae; Regulatory networks; Transcription factors; Virulence.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Architecture of genome-wide transcriptional regulatory network reveals dynamic functions and evolutionary trajectories in Pseudomonas syringae.Elife. 2025 Mar 31;13:RP96172. doi: 10.7554/eLife.96172. Elife. 2025. PMID: 40162990 Free PMC article.
-
Regulation of type III secretion system in Pseudomonas syringae.Environ Microbiol. 2019 Dec;21(12):4465-4477. doi: 10.1111/1462-2920.14779. Epub 2019 Aug 27. Environ Microbiol. 2019. PMID: 31408268 Review.
-
The transcriptional regulators of virulence for Pseudomonas aeruginosa: Therapeutic opportunity and preventive potential of its clinical infections.Genes Dis. 2022 Oct 1;10(5):2049-2063. doi: 10.1016/j.gendis.2022.09.009. eCollection 2023 Sep. Genes Dis. 2022. PMID: 37492705 Free PMC article. Review.
-
Integrated regulatory network in Pseudomonas syringae reveals dynamics of virulence.Cell Rep. 2021 Mar 30;34(13):108920. doi: 10.1016/j.celrep.2021.108920. Cell Rep. 2021. PMID: 33789108
-
Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals.Microorganisms. 2021 Jun 5;9(6):1227. doi: 10.3390/microorganisms9061227. Microorganisms. 2021. PMID: 34198761 Free PMC article. Review.
Cited by
-
VimR is a conserved master transcription regulator controlling virulence and metabolism in Pseudomonas.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf493. doi: 10.1093/nar/gkaf493. Nucleic Acids Res. 2025. PMID: 40498071 Free PMC article.
-
DNA methylome regulates virulence and metabolism in Pseudomonas syringae.Elife. 2025 Feb 24;13:RP96290. doi: 10.7554/eLife.96290. Elife. 2025. PMID: 39992965 Free PMC article.
-
From freezing to functioning: cellular strategies of cold-adapted bacteria for surviving in extreme environments.Arch Microbiol. 2024 Jun 28;206(7):329. doi: 10.1007/s00203-024-04058-5. Arch Microbiol. 2024. PMID: 38940837 Review.
-
Architecture of genome-wide transcriptional regulatory network reveals dynamic functions and evolutionary trajectories in Pseudomonas syringae.Elife. 2025 Mar 31;13:RP96172. doi: 10.7554/eLife.96172. Elife. 2025. PMID: 40162990 Free PMC article.
-
The Role of Heat Shock Protein (Hsp) Chaperones in Environmental Stress Adaptation and Virulence of Plant Pathogenic Bacteria.Int J Mol Sci. 2025 Jan 9;26(2):528. doi: 10.3390/ijms26020528. Int J Mol Sci. 2025. PMID: 39859244 Free PMC article. Review.
References
-
- Ichinose Y., Taguchi F., Mukaihara T. Pathogenicity and virulence factors of Pseudomonas syringae. J Gen Plant Pathol. 2013;79(5):285–296.
-
- Taguchi F., Ichinose Y. Role of type IV Pili in virulence of Pseudomonas syringae pv. tabaci 6605: correlation of motility, multidrug resistance, and HR-inducing activity on a nonhost plant. Mol Plant Microbe In. 2011;24(9):1001–1011. - PubMed
-
- Kanda E., Tatsuta T., Suzuki T., Taguchi F., Naito K., Inagaki Y., et al. Two flagellar stators and their roles in motility and virulence in Pseudomonas syringae pv. tabaci 6605. Mol Genet Genomics. 2011;285(2):163–174. - PubMed
-
- Lee C.A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5(4):148–156. - PubMed
-
- Nomura K., Melotto M., He S.Y. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8(4):361–368. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
