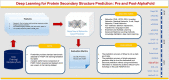
Deep learning for protein secondary structure prediction: Pre and post-AlphaFold
- PMID: 36420164
- PMCID: PMC9678802
- DOI: 10.1016/j.csbj.2022.11.012
Deep learning for protein secondary structure prediction: Pre and post-AlphaFold
Abstract
This paper aims to provide a comprehensive review of the trends and challenges of deep neural networks for protein secondary structure prediction (PSSP). In recent years, deep neural networks have become the primary method for protein secondary structure prediction. Previous studies showed that deep neural networks had uplifted the accuracy of three-state secondary structure prediction to more than 80%. Favored deep learning methods, such as convolutional neural networks, recurrent neural networks, inception networks, and graph neural networks, have been implemented in protein secondary structure prediction. Methods adapted from natural language processing (NLP) and computer vision are also employed, including attention mechanism, ResNet, and U-shape networks. In the post-AlphaFold era, PSSP studies focus on different objectives, such as enhancing the quality of evolutionary information and exploiting protein language models as the PSSP input. The recent trend to utilize pre-trained language models as input features for secondary structure prediction provides a new direction for PSSP studies. Moreover, the state-of-the-art accuracy achieved by previous PSSP models is still below its theoretical limit. There are still rooms for improvement to be made in the field.
Keywords: Deep neural networks; Machine learning; Prediction accuracy; Protein; Protein secondary structure prediction.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Protein secondary structure prediction improved by recurrent neural networks integrated with two-dimensional convolutional neural networks.J Bioinform Comput Biol. 2018 Oct;16(5):1850021. doi: 10.1142/S021972001850021X. J Bioinform Comput Biol. 2018. PMID: 30419785
-
ILMCNet: A Deep Neural Network Model That Uses PLM to Process Features and Employs CRF to Predict Protein Secondary Structure.Genes (Basel). 2024 Oct 21;15(10):1350. doi: 10.3390/genes15101350. Genes (Basel). 2024. PMID: 39457474 Free PMC article.
-
PSSP-MFFNet: A Multifeature Fusion Network for Protein Secondary Structure Prediction.ACS Omega. 2024 Jan 25;9(5):5985-5994. doi: 10.1021/acsomega.3c10230. eCollection 2024 Feb 6. ACS Omega. 2024. PMID: 38343972 Free PMC article.
-
Deep Learning for Natural Language Processing in Radiology-Fundamentals and a Systematic Review.J Am Coll Radiol. 2020 May;17(5):639-648. doi: 10.1016/j.jacr.2019.12.026. Epub 2020 Jan 28. J Am Coll Radiol. 2020. PMID: 32004480
-
Protein secondary structure prediction using neural networks and deep learning: A review.Comput Biol Chem. 2019 Aug;81:1-8. doi: 10.1016/j.compbiolchem.2019.107093. Epub 2019 Aug 12. Comput Biol Chem. 2019. PMID: 31442779 Review.
Cited by
-
Revealing protein sequence organization via contiguous hydrophobicity with the blobulator toolkit.bioRxiv [Preprint]. 2025 Mar 18:2024.01.15.575761. doi: 10.1101/2024.01.15.575761. bioRxiv. 2025. PMID: 38293114 Free PMC article. Preprint.
-
Advancements in one-dimensional protein structure prediction using machine learning and deep learning.Comput Struct Biotechnol J. 2025 Apr 3;27:1416-1430. doi: 10.1016/j.csbj.2025.04.005. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40242292 Free PMC article. Review.
-
ToxDL 2.0: Protein toxicity prediction using a pretrained language model and graph neural networks.Comput Struct Biotechnol J. 2025 Apr 2;27:1538-1549. doi: 10.1016/j.csbj.2025.04.002. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40276117 Free PMC article.
-
DeepPredict: a state-of-the-art web server for protein secondary structure and relative solvent accessibility prediction.Front Bioinform. 2025 Jun 6;5:1607402. doi: 10.3389/fbinf.2025.1607402. eCollection 2025. Front Bioinform. 2025. PMID: 40546733 Free PMC article.
-
Recent Advances in Computational Prediction of Secondary and Supersecondary Structures from Protein Sequences.Methods Mol Biol. 2025;2870:1-19. doi: 10.1007/978-1-0716-4213-9_1. Methods Mol Biol. 2025. PMID: 39543027 Review.
References
-
- Breda A., Valadares N.F., de Souza O.N., Garratt R.C. In: Gruber A., Durham A.M., Huynh C., del Portillo H.A., editors. Ch. A06. National Center for Biotechnology Information (US); Bethesda (MD): 2008. Protein structure, modelling and applications; pp. 137–170. (Bioinformatics in tropical disease research: a practical and case-study approach).
-
- Branden C.I., Tooze J. Introduction to Protein Structure. Garland Sci. 2012 doi: 10.1201/9781136969898. - DOI
-
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
