Distinction of early complement classical and lectin pathway activation via quantification of C1s/C1-INH and MASP-1/C1-INH complexes using novel ELISAs
- PMID: 36420270
- PMCID: PMC9677118
- DOI: 10.3389/fimmu.2022.1039765
Distinction of early complement classical and lectin pathway activation via quantification of C1s/C1-INH and MASP-1/C1-INH complexes using novel ELISAs
Erratum in
-
Corrigendum: Distinction of early complement classical and lectin pathway activation via quantification of C1s/C1-INH and MASP-1/C1-INH complexes using novel ELISAs.Front Immunol. 2025 Feb 12;16:1561850. doi: 10.3389/fimmu.2025.1561850. eCollection 2025. Front Immunol. 2025. PMID: 40013135 Free PMC article.
Abstract
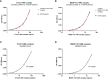
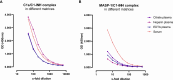
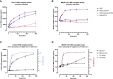
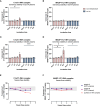
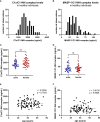
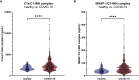
The most commonly used markers to assess complement activation are split products that are produced through activation of all three pathways and are located downstream of C3. In contrast, C4d derives from the cleavage of C4 and indicates either classical (CP) or lectin pathway (LP) activation. Although C4d is perfectly able to distinguish between CP/LP and alternative pathway (AP) activation, no well-established markers are available to differentiate between early CP and LP activation. Active enzymes of both pathways (C1s/C1r for the CP, MASP-1/MASP-2 for the LP) are regulated by C1 esterase inhibitor (C1-INH) through the formation of covalent complexes. Aim of this study was to develop validated immunoassays detecting C1s/C1-INH and MASP-1/C1-INH complex levels. Measurement of the complexes reveals information about the involvement of the respective pathways in complement-mediated diseases. Two sandwich ELISAs detecting C1s/C1-INH and MASP-1/C1-INH complex were developed and tested thoroughly, and it was investigated whether C1s/C1-INH and MASP-1/C1-INH complexes could serve as markers for either early CP or LP activation. In addition, a reference range for these complexes in healthy adults was defined, and the assays were clinically validated utilizing samples of 414 COVID-19 patients and 96 healthy controls. The immunoassays can reliably measure C1s/C1-INH and MASP-1/C1-INH complex concentrations in EDTA plasma from healthy and diseased individuals. Both complex levels are increased in serum when activated with zymosan, making them suitable markers for early classical and early lectin pathway activation. Furthermore, measurements of C1-INH complexes in 96 healthy adults showed normally distributed C1s/C1-INH complex levels with a physiological concentration of 1846 ± 1060 ng/mL (mean ± 2SD) and right-skewed distribution of MASP-1/C1-INH complex levels with a median concentration of 36.9 (13.18 - 87.89) ng/mL (2.5-97.5 percentile range), while levels of both complexes were increased in COVID-19 patients (p<0.0001). The newly developed assays measure C1-INH complex levels in an accurate way. C1s/C1-INH and MASP-1/C1-INH complexes are suitable markers to assess early classical and lectin pathway activation. An initial reference range was set and first studies showed that these markers have added value for investigating and unraveling complement activation in human disease.
Keywords: C1-INH complexes; C1s/C1-INH complex; MASP-1/C1-INH complex; assay development and validation; classical pathway activation; early complement activation; lectin pathway activation.
Copyright © 2022 Hurler, Toonen, Kajdácsi, van Bree, Brandwijk, de Bruin, Lyons, Bergamaschi, Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) COVID BioResource Collaboration, Sinkovits, Cervenak, Würzner and Prohászka.
Conflict of interest statement
ET, BB, RB and WB are employees of Hycult Biotech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

