The Aer2 chemoreceptor from Vibrio vulnificus is a tri-PAS-heme oxygen sensor
- PMID: 36420630
- PMCID: PMC10107281
- DOI: 10.1111/mmi.15007
The Aer2 chemoreceptor from Vibrio vulnificus is a tri-PAS-heme oxygen sensor
Abstract
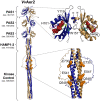
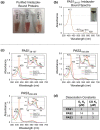
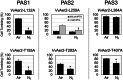
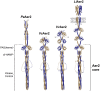
The marine pathogen Vibrio vulnificus senses and responds to environmental stimuli via two chemosensory systems and 42-53 chemoreceptors. Here, we present an analysis of the V. vulnificus Aer2 chemoreceptor, VvAer2, which is the first V. vulnificus chemoreceptor to be characterized. VvAer2 is related to the Aer2 receptors of other gammaproteobacteria, but uncharacteristically contains three PAS domains (PAS1-3), rather than one or two. Using an E. coli chemotaxis hijack assay, we determined that VvAer2, like other Aer2 receptors, senses and responds to O2 . All three VvAer2 PAS domains bound pentacoordinate b-type heme and exhibited similar O2 affinities. PAS2 and PAS3 both stabilized O2 via conserved Iβ-Trp residues, but PAS1, which was easily oxidized in vitro, was unaffected by Iβ-Trp replacement. Our results support a model in which PAS1 is largely dispensable for O2 -mediated signaling, whereas PAS2 modulates PAS3 signaling, and PAS3 signals to the downstream domains. Each PAS domain appeared to be positionally optimized, because PAS swapping caused altered signaling properties, and neither PAS1 nor PAS2 could replace PAS3. Our findings strengthen previous conclusions that Aer2 receptors are O2 sensors, but with distinct N-terminal domain arrangements that facilitate, modulate and tune responses based on environmental signals.
Keywords: Vibrio vulnificus; PAS domain; chemoreceptor; oxygen sensing; signal transduction.
© 2022 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no known conflict of interest.
Figures






References
-
- Baker‐Austin, C. , Oliver, J.D. , Alam, M. , Ali, A. , Waldor, M.K. , Qadri, F. et al. (2018) Vibrio spp. infections. Nature Reviews. Disease Primers, 4, 1–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

