The UCSC Genome Browser database: 2023 update
- PMID: 36420891
- PMCID: PMC9825520
- DOI: 10.1093/nar/gkac1072
The UCSC Genome Browser database: 2023 update
Abstract
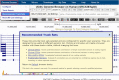
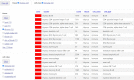
The UCSC Genome Browser (https://genome.ucsc.edu) is an omics data consolidator, graphical viewer, and general bioinformatics resource that continues to serve the community as it enters its 23rd year. This year has seen an emphasis in clinical data, with new tracks and an expanded Recommended Track Sets feature on hg38 as well as the addition of a single cell track group. SARS-CoV-2 continues to remain a focus, with regular annotation updates to the browser and continued curation of our phylogenetic sequence placing tool, hgPhyloPlace, whose tree has now reached over 12M sequences. Our GenArk resource has also grown, offering over 2500 hubs and a system for users to request any absent assemblies. We have expanded our bigBarChart display type and created new ways to visualize data via bigRmsk and dynseq display. Displaying custom annotations is now easier due to our chromAlias system which eliminates the requirement for renaming sequence names to the UCSC standard. Users involved in data generation may also be interested in our new tools and trackDb settings which facilitate the creation and display of their custom annotations.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures