GENCODE: reference annotation for the human and mouse genomes in 2023
- PMID: 36420896
- PMCID: PMC9825462
- DOI: 10.1093/nar/gkac1071
GENCODE: reference annotation for the human and mouse genomes in 2023
Abstract
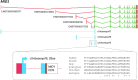
GENCODE produces high quality gene and transcript annotation for the human and mouse genomes. All GENCODE annotation is supported by experimental data and serves as a reference for genome biology and clinical genomics. The GENCODE consortium generates targeted experimental data, develops bioinformatic tools and carries out analyses that, along with externally produced data and methods, support the identification and annotation of transcript structures and the determination of their function. Here, we present an update on the annotation of human and mouse genes, including developments in the tools, data, analyses and major collaborations which underpin this progress. For example, we report the creation of a set of non-canonical ORFs identified in GENCODE transcripts, the LRGASP collaboration to assess the use of long transcriptomic data to build transcript models, the progress in collaborations with RefSeq and UniProt to increase convergence in the annotation of human and mouse protein-coding genes, the propagation of GENCODE across the human pan-genome and the development of new tools to support annotation of regulatory features by GENCODE. Our annotation is accessible via Ensembl, the UCSC Genome Browser and https://www.gencodegenes.org.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Frankish A., Uszczynska B., Ritchie G.R.S., Gonzalez J.M., Pervouchine D., Petryszak R., Mudge J.M., Fonseca N., Brazma A., Guigo R.et al. .. Comparison of GENCODE and refseq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics. 2015; 16(Suppl. 8):S2. - PMC - PubMed

